
haobo wang
@wwwhbptwork for protein design at https://t.co/ODnlN6OGef, a previous postdoc with @sokrypton at Harvard
Similar User

@KathyYWei1

@minkbaek

@JustasDauparas

@proteinrosh

@jueseph

@BrianHie

@befcorreia

@awakenkunkun

@ZiboChen

@namrata_anand2

@FurmanLab

@Chica_lab

@alexechu_

@naterbennett0

@CasperGoverde
🤯 Congratulations David Baker @UWproteindesign , @demishassabis and John Jumper!!
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”

1/X Pallatom is an end-to-end all-atom protein generation model that directly learns the distribution of atomic coordinates. We believe optimizing the P(all-atom) may open a new way for de novo protein design. code(ing):github.com/levinthal/Pall… bioxiv:biorxiv.org/content/10.110…
OMG 😱 finally a protein language model that captures coevolution at protein-protein interface(s)!
7/ In particular, we showcase gLM2's ability to directly learn coevolutionary signal in protein-protein interfaces with no supervision! The learned contact maps can be extracted using @ZhidianZ et al's categorial Jacobian method.

Excited to share the work with @ZhidianZ @HWaymentSteele @garykbrixi @wwwhbpt @matteodp @DorotheeKern biorxiv.org/content/10.110… (1/7)
I'm excited to share that I'll be joining @MITBiology as an Asst Prof. in Jan 2024! Come join us! 🤓🧪🖥️🧬

Now everyone can be a protein designer! 😂
The code is available both to download from GitHub, and also, thanks to the wonderful @sokrypton, as a Colab Notebook. github.com/RosettaCommons… colab.research.google.com/github/sokrypt…
Today we're making RF Diffusion, our guided diffusion model for protein design with potential applications in medicine, vaccines & advanced materials, free to use. The software has proven much faster and more capable than prior protein design tools. bakerlab.org/2023/03/30/rf-…
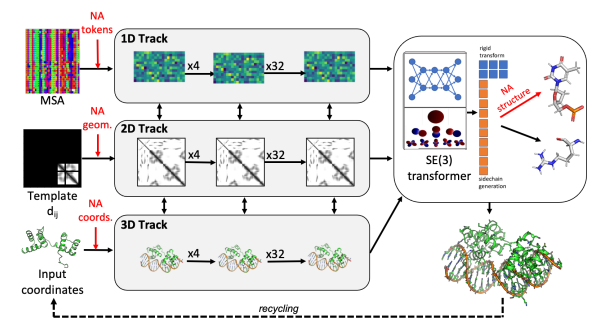
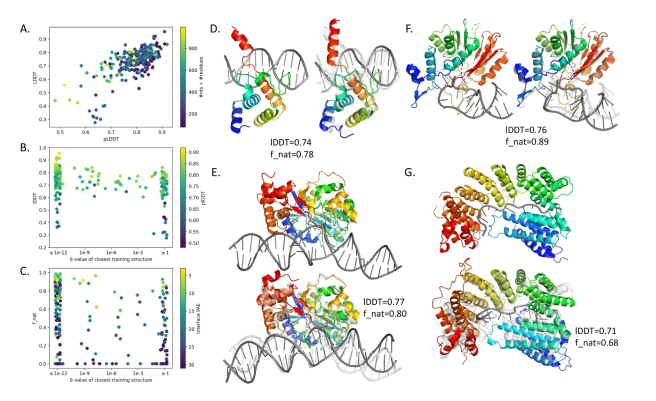
A single neural network that predicts for protein-DNA complex structures, protein-RNA complex structures, and RNA tertiary structures. @minkbaek Ryan McHugh, Ivan Anishchenko, David Baker, Frank DiMaio @UWproteindesign biorxiv.org/content/10.110…
Using AF2 to predict minibinder-target complexes increases experimental success rate 10x over filtering by non-DL in silico metrics. This + ProteinMPNN has totally changed binder design in Baker lab.
1/7 Improving de novo Protein Binder Design with Deep Learning (biorxiv.org/content/10.110…) We show that AF2 is an effective predictor of whether a de novo designed miniprotein will bind to the intended target or not.
ProteinMPNN designed binders work experimentally! Using ProteinMPNN to design miniprotein binders is 6-fold more computationally efficient. Also, the probability that a miniprotein binds the target as designed increases 10-fold when using AlphaFold (or RoseTTAFold) as filters!
1/7 Improving de novo Protein Binder Design with Deep Learning (biorxiv.org/content/10.110…) We show that AF2 is an effective predictor of whether a de novo designed miniprotein will bind to the intended target or not.
Code should be publicly available now! Thanks @sokrypton for making a quick demo on google colab: github.com/dauparas/Prote…
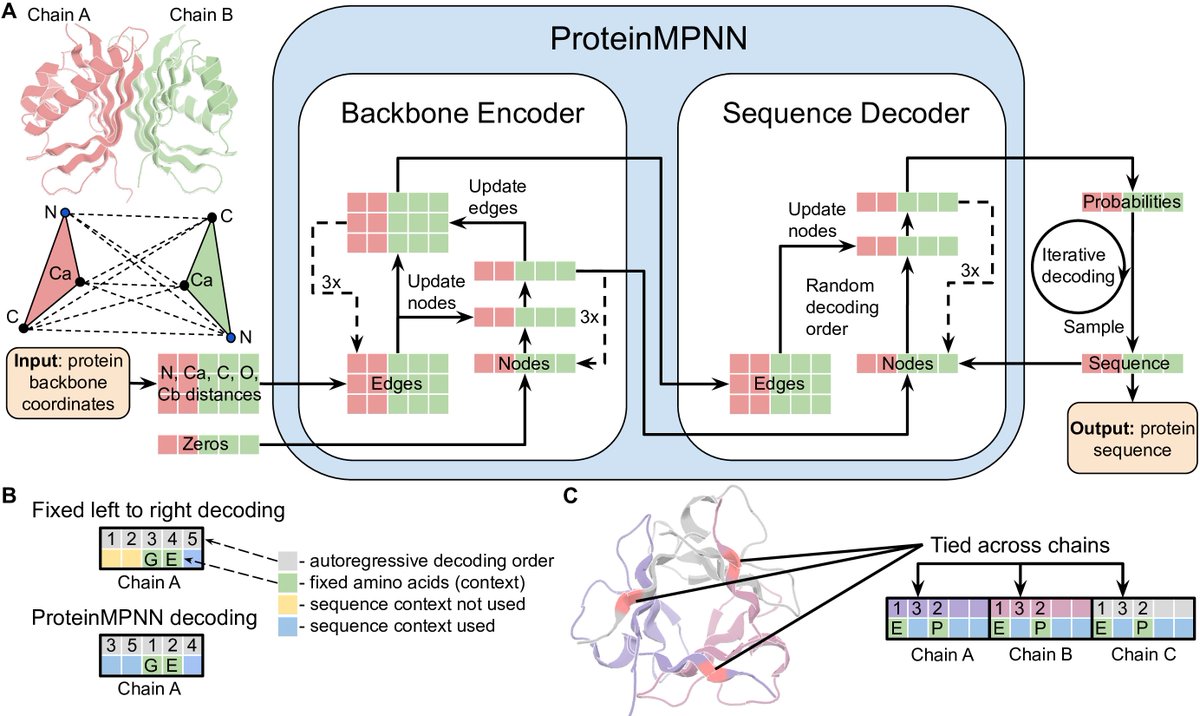
1/5 Robust deep learning based protein sequence design using ProteinMPNN biorxiv.org/content/10.110… The high rate of experimental success and applicability to almost any protein sequence design problem has made it the standard approach at the @UWproteindesign github.com/dauparas/Prote…
Excited to see colabfold published! nature.com/articles/s4159… Special thanks to @thesteinegger and @milot_mirdita (for MMseqs2) without whom I would have never considered to preprint let alone attempt to publish our notebook! (1/3)
Foldseek got a 3D structure visualization using NGL thanks to my postdoc @clmgilchrist and @milot_mirdita We generate missing atoms using pulchra and superpose aligned sequences using TMalign in the browser using #WebAssembly 🌐 search.foldseek.com 📄 biorxiv.org/content/10.110…
Here’s what we learned from inverse folding on millions of #AlphaFold structures. Exciting time to bring a 800x new scale to #proteindesign. ESM-IF1 more accurately designs sequences to fold into desired structure, also unlocking new design capabilities. biorxiv.org/content/10.110…
We frequently rely on google scholar to assess academic impact. However, most people don't realize you can edit the h-index on your profile! Check the demo below to see how. This is especially important for people like me, whose h-index doesn't reflect my actual academic impact
United States Trends
- 1. Good Sunday 60,4 B posts
- 2. #chillguy 27,8 B posts
- 3. #sundayvibes 6.256 posts
- 4. Chuck Woolery 9.040 posts
- 5. #askzb N/A
- 6. #SundayMotivation 2.799 posts
- 7. #SundayFunday 1.787 posts
- 8. Wheel of Fortune 4.255 posts
- 9. Love Connection 5.564 posts
- 10. Blessed Sunday 15,3 B posts
- 11. Lakers 42 B posts
- 12. F-35 23,4 B posts
- 13. Bama 39,3 B posts
- 14. Auburn 36,8 B posts
- 15. Lord's Day 2.655 posts
- 16. King of the Universe 1.733 posts
- 17. Christ Jesus 50,5 B posts
- 18. Anne 55 B posts
- 19. Max Verstappen 146 B posts
- 20. Pant 74,5 B posts
Who to follow
-
 Kathy Y. Wei, Ph.D.
Kathy Y. Wei, Ph.D.
@KathyYWei1 -
 Minkyung Baek
Minkyung Baek
@minkbaek -
 Justas Dauparas
Justas Dauparas
@JustasDauparas -
 Roshan Rao
Roshan Rao
@proteinrosh -
 Jue Wang
Jue Wang
@jueseph -
 Brian Hie
Brian Hie
@BrianHie -
 Bruno Correia
Bruno Correia
@befcorreia -
 Weikun.Wu
Weikun.Wu
@awakenkunkun -
 Zibo Chen
Zibo Chen
@ZiboChen -
 Namrata Anand
Namrata Anand
@namrata_anand2 -
 Ora Furman Lab 🎗
Ora Furman Lab 🎗
@FurmanLab -
 Roberto Chica Lab
Roberto Chica Lab
@Chica_lab -
 Alex Chu
Alex Chu
@alexechu_ -
 Nate Bennett
Nate Bennett
@naterbennett0 -
 Casper Goverde
Casper Goverde
@CasperGoverde
Something went wrong.
Something went wrong.









































































































