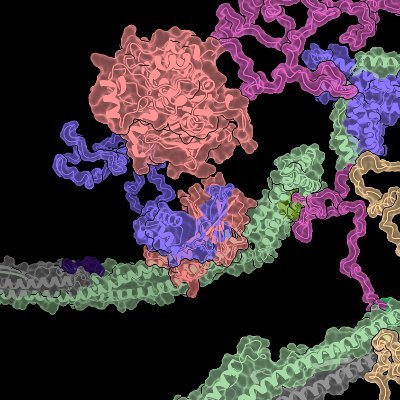
Three months ago, we launched #ribonanza, hoping to accelerate progress in #deeplearning RNA structure prediction. A dual crowdsourcing effort involving chemical mapping data on ~2.1M RNA sequences @eternagame and @kaggle kaggle.com/c/stanford-rib… Results are in! 1/N
Model whose weights I dream about seeing someday: AlphaFold-latest, Chroma
Protein model I dislike: EvoDiff-MSA Model I begrudgingly respect: AlphaFold Model I think is overrated: RFdiffusion model I think is underrated: Genie Model I like: CARP Model I love: ProtPardelle Model I dream of designing proteins with: EvoDiff-seq
For protein modeling tools. Tool that ... I dislike: Rosetta I begrudgingly respect: BLASTp I think is overrated: PyMOL I think is underrated: ChimeraX I like: Foldseek I love: SeqKit I dream that exist one day: protein emoji 💔 pic of the eternally compilation of Rosetta🫠😮💨

Protein model I dislike: EvoDiff-MSA Model I begrudgingly respect: AlphaFold Model I think is overrated: RFdiffusion model I think is underrated: Genie Model I like: CARP Model I love: ProtPardelle Model I dream of designing proteins with: EvoDiff-seq
I spent a lot of time trying this when I first started at MSR, but I could never get it to work. The keys: - Generate many possible trees instead of just using the ancestral nodes from one. - Consider indels. biorxiv.org/content/10.110…
DeepLocPro: Use ESM-2 embeddings to predict prokaryotic protein subcellular localization. I'm kinda surprised that this is the first deep learning method for prokaryotic localization and that the SotA is from 2010! @felixgteufel

Different kinds of external knowledge are valuable to build the relationship between the seen and unseen classes in a zero shot setting, including text, attribute, KG, rule & ontology, according to their characteristics, expressivity and the semantic encoding approaches.

How to do influential research: a few lessons learned by Xiaodong He. Great summary!

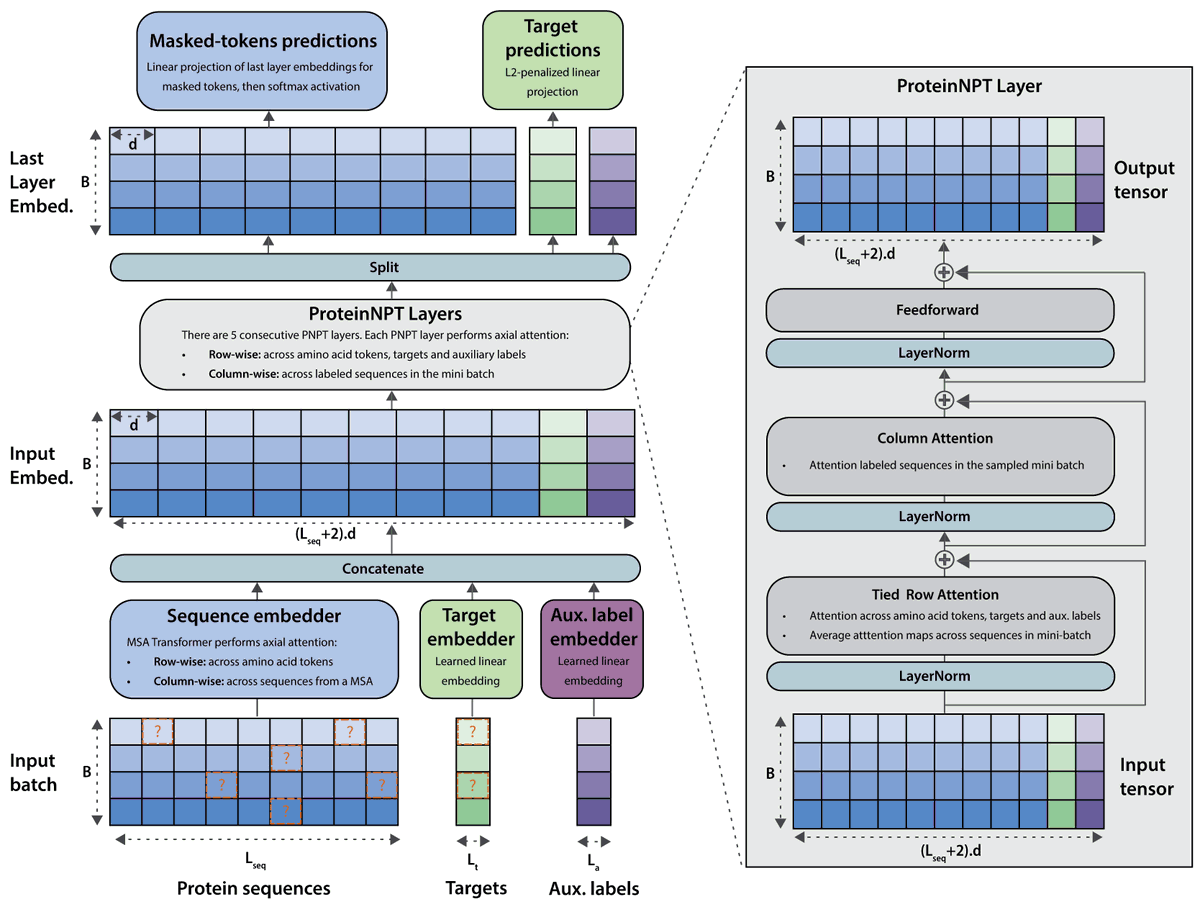
Train a neural network to reconstruct both masked amino acids and masked labels while allowing attention between sequences in order to improve both fitness prediction and sequence generation. biorxiv.org/content/10.110… @NotinPascal @ruben_weitzman @deboramarks @yaringal

📢Very pleased to be presenting our ProteinNPT paper at NeurIPS on Tuesday!📢We introduce a novel semi-supervised conditional pseudo-generative model for fitness prediction and iterative protein redesign biorxiv.org/content/10.110… #NeurIPS2023 #GenBio #ProteinDesign
One advice from my postdoc advisor has always resonated with me: 'You should only do things only you can do.' It's obviously a lofty standard Ill never be able to reach. But after all, if something can be easily done or scooped by others, what unique value are we adding ??
Compbio community has always been ahead of the times. Early adoption of preprints, sharing unpublished work early etc. Those who continue to be paranoid & secretive lose out. Getting feedback & collaborating early is the best way to improve research.
Introducing Gemini 1.0, our most capable and general AI model yet. Built natively to be multimodal, it’s the first step in our Gemini-era of models. Gemini is optimized in three sizes - Ultra, Pro, and Nano Gemini Ultra’s performance exceeds current state-of-the-art results on…

When comparing two models, a common reference point of compute is often used. If you trained a 7b model with 3x the number of tokens/compute to beat a 13b model, did you really beat it? Probably not. 😶 Here's a paper we wrote in 2021 (arxiv.org/abs/2110.12894) that I still…
I am excited to present this work, result of a 4-year big collaborative project: arxiv.org/abs/2310.18278 #MachineLearning a transferable bottom-up protein force field, trained on force data from over all-atom MD simulations, using physical priors and graph neural networks.🧵⬇️
Large language models (LLMs) can make small talk with you. But can they navigate more difficult real-life social scenarios? 👋 Meet SOTOPIA sotopia.world - our new multi-agent social environment from CMU that answers this question (collab w/ @nlpxuhui et al.). 🤖
Tuna: Instruction Tuning using Feedback from Large Language Models paper page: huggingface.co/papers/2310.13… Instruction tuning of open-source large language models (LLMs) like LLaMA, using direct outputs from more powerful LLMs such as Instruct-GPT and GPT-4, has proven to be a…

1.17B proteins from 26,931 metagenomes with no similarity to anything in reference genomes. Cluster into 106,198 novel clusters with more than 100 members and predict structures! @g_pavlopoulos @BSRC_Fleming @kyrpides @jgi @SiruiLiu_ @sokrypton nature.com/articles/s4158…



I wrote about how neural network hallucinations can be used to design novel proteins. Excited about a future where we can rapidly design smart therapeutics that disrupt disease. liambai.com/protein-halluc…
"The Protein Engineering Tournament: An Open Science Benchmark for Protein Modeling and Design" "A fully-remote, biennial competition for the development and benchmarking of computational methods in protein engineering" Hope you'll all participate! arxiv.org/abs/2309.09955

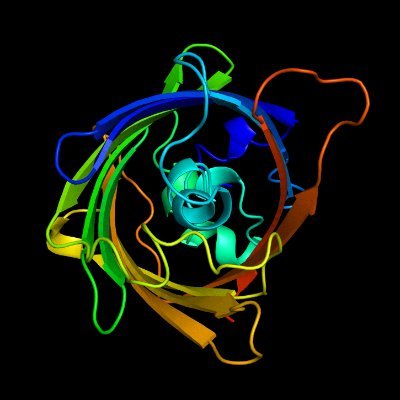
"Accurately identifying nucleic-acid-binding sites through geometric graph learning on language model predicted structures" has been updated Graph-based approach that works on ESMFold-predicted structures biorxiv.org/content/10.110… github.com/biomed-AI/nucl…

"Alignment-based protein mutational landscape prediction: doing more with less" has been updated Performance exceeds HMMs and is comparable to general PLMs and family-specific models. biorxiv.org/content/10.110… lcqb.upmc.fr/GEMME/Home.html

United States Trends
- 1. Thanksgiving 846 B posts
- 2. Druski 34,9 B posts
- 3. UConn 11,7 B posts
- 4. Kevin Hart 21,1 B posts
- 5. Wiggins 4.911 posts
- 6. Dylan Harper 4.784 posts
- 7. Dayton 5.403 posts
- 8. #RHOSLC 6.999 posts
- 9. Zuckerberg 63,7 B posts
- 10. Shai 7.791 posts
- 11. Friday Night Lights 19,9 B posts
- 12. Vindman 74,5 B posts
- 13. Pat Spencer 1.385 posts
- 14. 0-3 in Maui N/A
- 15. Jalen Williams 2.255 posts
- 16. Tyrese Martin 2.216 posts
- 17. #MSIxSTALKER2 7.418 posts
- 18. Enoch Cheeks N/A
- 19. #BillboardIsOverParty 161 B posts
- 20. Adin Hill N/A
Something went wrong.
Something went wrong.