Similar User

@sedlazeck

@StevenSalzberg1

@alexisjbattle

@khmiga

@The__Taybor

@mitenjain

@GenomeInABottle

@mike_schatz

@Hasindu2008

@MishaKolmogorov

@mattloose

@PradeepMadapura

@JonathanGoeke

@aquaskyline

@wouter_decoster
Interested in a #postdoc position to develop new sequencing methods? Come work with us! Work includes but is not limited to epigenetics, RNA mods, and single-cell/spatial transcriptomics. docs.google.com/document/d/1ZM…
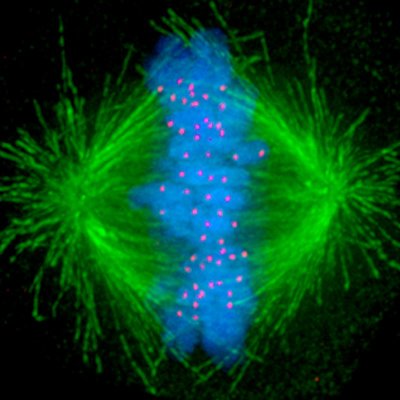
It's my pleasure to present the next big preprint from SheqLab! An exciting application of our O-MAP platform that I hope will transform the study of nuclear architecture. If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)

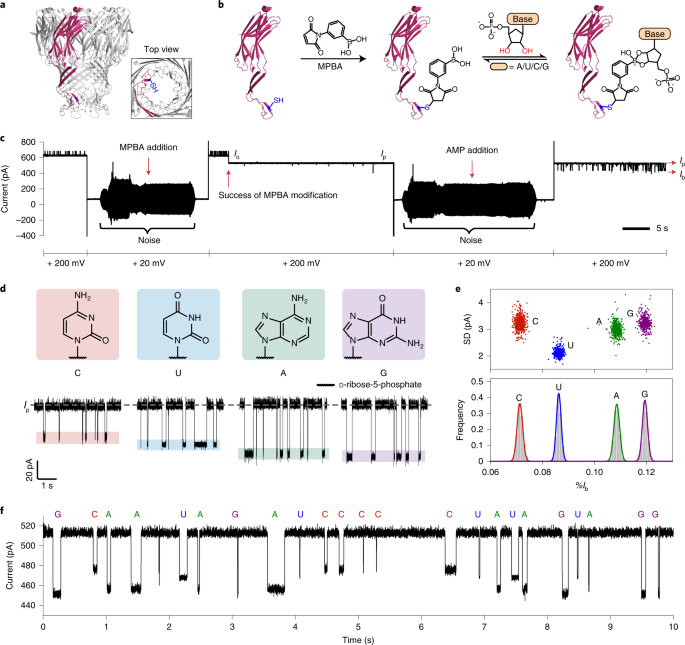
Our preprint is out! We hacked the @nanopore sequencer to read amino acids and PTMs along protein strands. This opens up the possibility for barcode sequencing at the protein level for highly multiplexed assays, PTM monitoring, and protein identification! biorxiv.org/content/10.110…
Beyond assembly: the increasing flexibility of single-molecule sequencing technology go.nature.com/3HSiBd4 #Review by @pwhook & @timp0 @JohnsHopkins @JHUBME
Check out the beta release of @alexsweeten's new tool Mod.Plot! We "mashed up" @mrvollger's StainedGlass visualization with minhashing to enable instant and interactive dotplotting. No alignment needed! Preprint of the method to follow in a couple months 📈
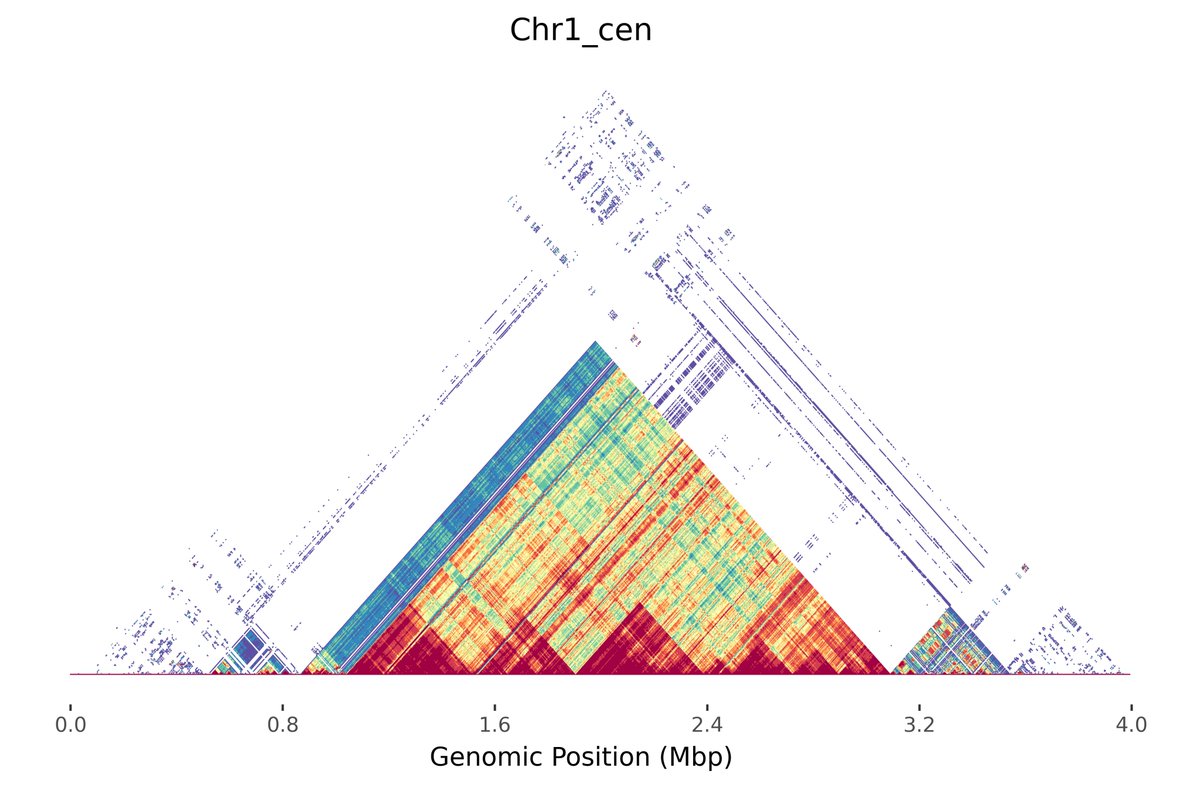
I'll be presenting Mod.Plot, a fast and fully interactive dot plot viewer, today at the @RECOMBseq poster session! github.com/marbl/ModDotPl…
I'm excited to see this out! @DongzeHe & I have been working on simpleaf for a while, and it's already become our daily driver & preferred interface for single-cell processing. It has also been important in easing integration of alevin-fry into @nf_core's scRNA-seq pipeline! 1/2
simpleaf: A simple, flexible, and scalable framework for single-cell transcriptomics data processing using alevin-fry biorxiv.org/cgi/content/sh… #bioRxiv
We look towards a future of nanopore protein sequencing when single-molecule meets single-cell. scPeptide-barcoding, full-length protein reads, and “near-pore” processing…
A Comment by @KeisukeMotone and @jeffnivala highlights the challenges of fully decoding amino acids and their modifications using protein nanopore sequencing technology, and discusses its unique potential for single-cell proteomics. nature.com/articles/s4159…

So thrilled the @lustgartenfdn is supporting this cutting-edge project to unlock the inherited genetic basis of pancreatic cancer using long-read sequencing. My co-leaders @mike_schatz @timp0 are true pioneers in this area. WE ARE HIRING!
When it comes to #pancreaticcancer, time is everything! 💜 @hopkinskimmel bit.ly/3FswcWo
These data provide invaluable information for future work to understand the mechanisms underlying formation and amplification of mitochondrial genome deletions and for using these variants as a metric of aging. Thank you to @timp0 for collaboration!
Using nanopore sequencing in human muscle, we quantify a higher frequency of mtDNA deletions than previously reported. The frequency correlates strongly with chronological age and is validated through use of an orthogonal quantification method (droplet digital PCR).
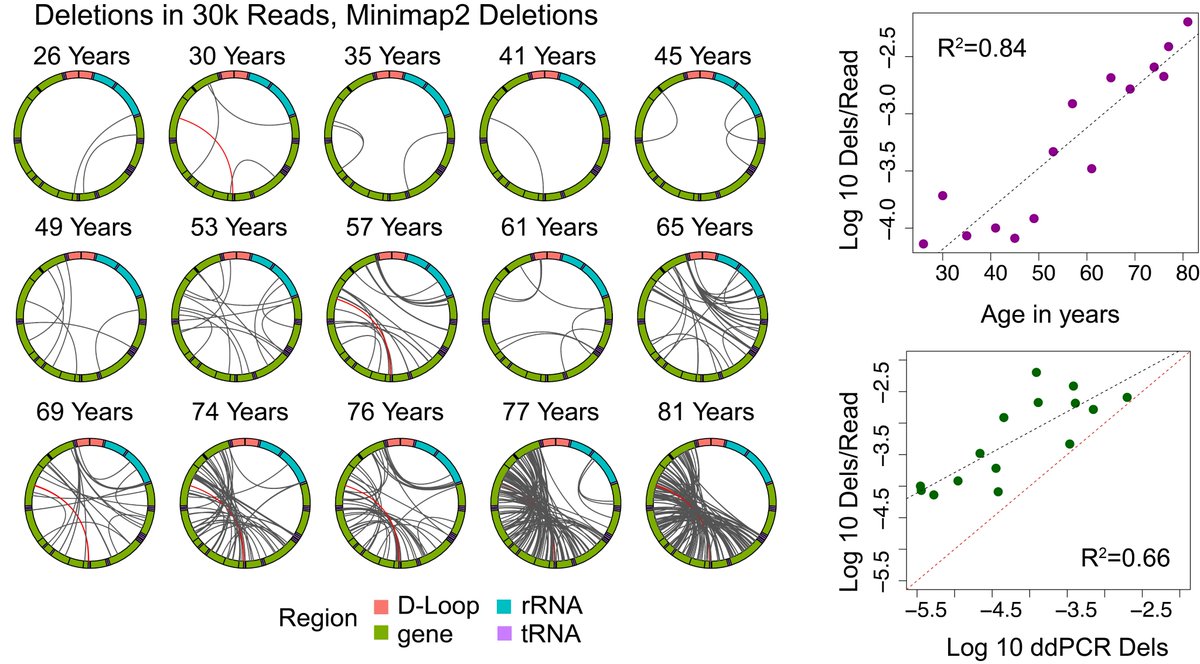
Excited to share our preprint using nanopore sequencing to map and quantify large deletion mutations in human mitochondrial DNA across the lifespan: biorxiv.org/content/10.110…. Highlights below:
We at @PacBio are finally presenting to the world, at long last, isoform information at #singlecell resolution - some details on the tech background for the kit and #Bioinformatics below 🧵
We’ve partnered with the @broadinstitute and @10xgenomics to launch the Multiplexed Arrays Sequencing (MAS-Seq) kit, an accurate and cost-effective long-read single cell #RNASeq solution. Learn more here: bit.ly/3MNGlR5 #PacBio #ASHG22
Congratulations @JustusKebschull, @JohnsHopkins recipient of a 2022 Klingenstein-Simons Fellowship Award in Neuroscience for the project “Mechanisms of evolutionary circuit expansion,” bit.ly/3LMrSUH @SimonsFdn

Long-read scRNA-seq is awesome for studying RNA isoforms. But @nanopore has relied on matched short-reads to identify cell barcodes and assign reads to cells. Pleased to share BLAZE, a new tool to identify cell barcodes from nanopore scRNA-seq. Short-reads not required. 1/4
Identification of cell barcodes from long-read single-cell RNA-seq with BLAZE biorxiv.org/cgi/content/sh… #bioRxiv
Excited to share our paper, out today in @Nature, where we characterize transcriptome variation in human tissues by long-read sequencing. Huge thanks to @g_dafni @beryl_bbc Garrett Garborcauskas @dgmacarthur and other collaborators. 🧵 nature.com/articles/s4158…
Genome editing + nanopore sequencing (DNA, RNA, and yes even protein sequencing too!) Oh my! Apply here to work with @chofski and I on an exciting new synbio venture👇🏼
📢New #synbio postdoc position in my lab on "Nanopore-based monitoring of yeast for bioprocess optimisation" with @jeffnivala Looking for someone obsessed by genome editing✂️ and nanopore sequencing🧬 (commercialisation interest a plus). Deadline 15th Aug shorturl.at/CGHP4


New content online: Identification of nucleoside monophosphates and their epigenetic modifications using an engineered nanopore dlvr.it/SV698g
Happy to share our latest work from @timp0 lab, modbamtools: Analysis of single-molecule epigenetic data for long-range profiling, heterogeneity, and clustering 👉manual and tutorial: rrazaghi.github.io/modbamtools/ 👉preprint: biorxiv.org/content/10.110… some of the tools to highlight: 1/
United States Trends
- 1. $ELONIA N/A
- 2. Mack Brown N/A
- 3. Johnie Cooks N/A
- 4. $CUTO 6.188 posts
- 5. #GivingTuesday 2.217 posts
- 6. Tariffs 184 B posts
- 7. Happy Thanksgiving 23,1 B posts
- 8. #tuesdayvibe 4.846 posts
- 9. #csm185 2.090 posts
- 10. #PumpRules N/A
- 11. Bill Gates 42,2 B posts
- 12. Shaq Barrett 1.194 posts
- 13. Canada 462 B posts
- 14. #TTPDTheAnthology 3.068 posts
- 15. Pitchfork 5.943 posts
- 16. Taco Tuesday 9.802 posts
- 17. Frederick 7.826 posts
- 18. Chanyeol 123 B posts
- 19. Alec Baldwin 6.713 posts
- 20. SNKRS 1.369 posts
Who to follow
-
 Fritz Sedlazeck
Fritz Sedlazeck
@sedlazeck -
Steven Salzberg 💙💛
@StevenSalzberg1 -
 Alexis Battle
Alexis Battle
@alexisjbattle -
 Karen Miga
Karen Miga
@khmiga -
 Clive G. Brown
Clive G. Brown
@The__Taybor -
 Miten Jain
Miten Jain
@mitenjain -
 Genome in a Bottle
Genome in a Bottle
@GenomeInABottle -
 Michael Schatz
Michael Schatz
@mike_schatz -
 Hasindu Gamaarachchi
Hasindu Gamaarachchi
@Hasindu2008 -
 Misha Kolmogorov
Misha Kolmogorov
@MishaKolmogorov -
 Matt Loose
Matt Loose
@mattloose -
 Pradeepa Madapura
Pradeepa Madapura
@PradeepMadapura -
 Jonathan Göke
Jonathan Göke
@JonathanGoeke -
 Ruibang Laurent Luo
Ruibang Laurent Luo
@aquaskyline -
 Wouter De Coster
Wouter De Coster
@wouter_decoster
Something went wrong.
Something went wrong.