Karen Miga
@khmigaAssociate Professor at UCSC; UC Santa Cruz Genomics Institute; Satellite DNA Biologist
Similar User

@glennis_logsdon

@HumanPangenome

@BenedictPaten

@marianattestad

@sedlazeck

@erikgarrison

@StevenSalzberg1

@GenomeInABottle

@alexisjbattle

@EichlerLab

@mike_schatz

@The__Taybor

@tobiasmarschal

@Magdoll

@infoecho
Accurate genome assemblies are key for research. DeepPolisher uses Pacbio HiFi alignments to improve base-level accuracy, cutting errors by half and reducing indel errors by over 70%. ✍🏼@miramastoras @kishwarshafin @BenedictPaten @MobinAsri Here's how... biorxiv.org/content/10.110…

Personalized pangenome preferences serve as crucial breakthroughs, furthering HPRC's mission to diversify genetic representation. @Nature highlights a more customized approach facilitating improvements in genetic diversity and reducing genotyping errors. nature.com/articles/s4159…
I am going on the faculty job market this year, so if your home department is starting a search, I'd love to hear about it. Thanks!
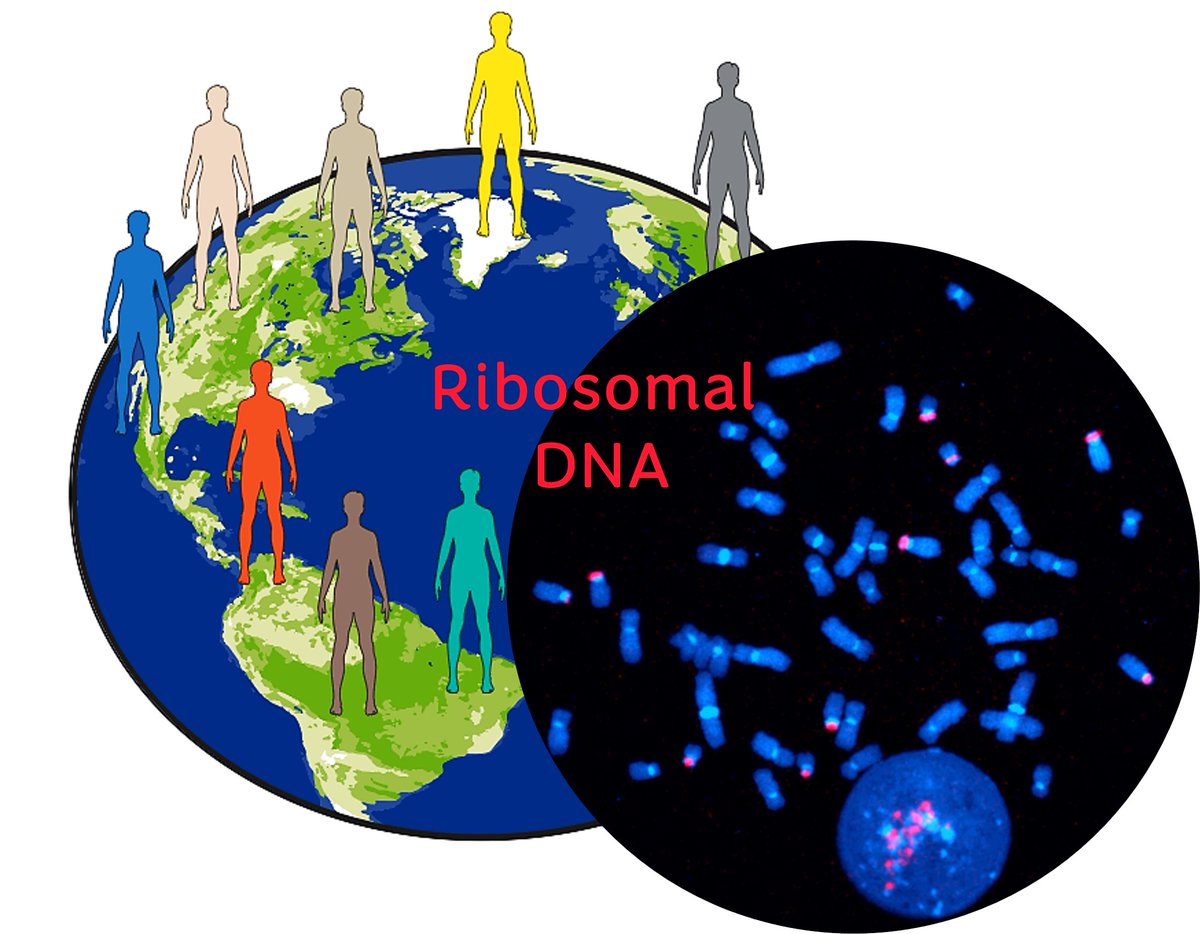
New preprint from our lab 🚨 We explored the variability in ribosomal DNA copy number and activity across humans of the world with @GertonJennifer @PaxtonKostos @AndresGuarahino @stevenjsolar @erikgarrison @aphillippy and others not on X biorxiv.org/content/10.110… 🧵
Genome sequencing is faster and cheaper than ever – but obstacles to broad adoption remain. Join @Google’s genomics lead @acarroll_ATG & @ucscgenomics’ @BenedictPaten for a live-streamed discussion on genomic analysis and equity: goo.gle/3ZmPyYO

Out today in Nature Methods, our first attempt make personalized pangenomes: rdcu.be/dTD3o In the future it will vital to subset pangenomes to just the relevant haplotypes for a compared sample. Work by @jltsiren, @27_matte, @par_esi, vg team, and many others.

Published today in @Nature, we describe an approach for single-molecule protein reading on @nanopore arrays. By utilizing ClpX unfoldase to ratchet proteins through a CsgG nanopore, we achieved single-amino-acid sensitivity. nature.com/articles/s4158…
A friendly reminder that today (August 1st) is the last day to register for the T2T-F2F! Hope you can join us (details below and reach out if you have any questions)
We welcome you to join us for the T2T Consortium Face-to-Face (T2T-F2F) Meeting in Santa Cruz, CA Sept 3-4. Open to all -- register before August 1st: sites.google.com/ucsc.edu/t2t-f…

If you’re interested in crafting a postdoc project at the intersection of biochemistry, structural biology, and circadian timekeeping that you can take with you to an independent position, reach out to me at partchlab.com. Let’s have some fun together!
Looking forward to rocking some fun new science in the lab as part of the @HHMINEWS community—many thanks to members of the @partchlab and our wonderful collaborators and colleagues for this incredible honor!
Our fast haplotagging paper is now published (nature.com/articles/s4146…) - see the earlier thread for a summary. Thanks to @kishwarshafin @AlexeyKolesni18 for leading the work and collaborators: @Johngorzynski, @gsneha261, @euanashley, @mitenjain, @khmiga, @BenedictPaten
Happy to share this paper which provides more detail about the process we use to assign haplotypes to long reads on-the-fly, which enabled us to speed up the DeepVariant release at v1.4. Implementation by Alexey Kolesnikov, Lead for collaboration, writing, figures @kishwarshafin
🧬Want to learn more about the Telomere-to-Telomere project and the folks involved in the T2T Consortium before the T2T-F2F meeting on Sept 3-4? 👇Watch the presentation @khmiga gave at the Genome Technology Forum held @jacksonlab in collab w/ @genome_gov youtu.be/t4kGX373EEQ
An exciting announcement from @illumina team -- of DRAGEN v4.3 includes 47 HPRC assemblies and is showing improvements over traditional mapping.
DRAGEN v4.3 by @illumina will compliment the work of #BaskinEngineering Assistant Prof. Karen Miga @khmiga, director of the Reference Production Center at the @HumanPangenome - "...a DRAGEN customer can now leverage all or a subset of the 47 HPRC samples." bit.ly/3XFpSG7
Groundbreaking work from @ProfDanielKim showcasing the potential of RNA liquid biopsies for early cancer detection #nanoporeconf

Nobel Prize Laureate, Carol Greider, kicked off this year's #nanoporeconf discussing the use of nanopore sequencing to reveal conservation of chromosome end-specific telomere lengths

United States Trends
- 1. Serrano 198 B posts
- 2. #TysonPaul 103 B posts
- 3. #NetflixFight 50,4 B posts
- 4. #netflixcrash 10,6 B posts
- 5. Shaq 11,7 B posts
- 6. #buffering 8.164 posts
- 7. Rosie Perez 10,1 B posts
- 8. Tori Kelly 3.522 posts
- 9. ROBBED 81,5 B posts
- 10. My Netflix 58,9 B posts
- 11. Ramos 65,3 B posts
- 12. Cedric 16,6 B posts
- 13. Jerry Jones 7.653 posts
- 14. Phil Collins 2.974 posts
- 15. #boxing 36,9 B posts
- 16. Christmas Day 13,8 B posts
- 17. WTF Netflix 13,6 B posts
- 18. Love is Blind 4.566 posts
- 19. The Hangover 3.734 posts
- 20. The Netflix 269 B posts
Who to follow
-
 Glennis Logsdon
Glennis Logsdon
@glennis_logsdon -
 Human Pangenome Reference Consortium
Human Pangenome Reference Consortium
@HumanPangenome -
 Benedict Paten
Benedict Paten
@BenedictPaten -
 Maria Nattestad
Maria Nattestad
@marianattestad -
 Fritz Sedlazeck
Fritz Sedlazeck
@sedlazeck -
 Erik Garrison
Erik Garrison
@erikgarrison -
Steven Salzberg 💙💛
@StevenSalzberg1 -
 Genome in a Bottle
Genome in a Bottle
@GenomeInABottle -
 Alexis Battle
Alexis Battle
@alexisjbattle -
 EichlerLab
EichlerLab
@EichlerLab -
 Michael Schatz
Michael Schatz
@mike_schatz -
 Clive G. Brown
Clive G. Brown
@The__Taybor -
 Tobias Marschall
Tobias Marschall
@tobiasmarschal -
 Liz Tseng
Liz Tseng
@Magdoll -
 Jason Chin
Jason Chin
@infoecho
Something went wrong.
Something went wrong.