David Porubsky
@PorubskyDavidBioinformatics, genomics & science art
Similar User

@glennis_logsdon

@EichlerLab

@mrvollger

@stergachislab

@HumanPangenome

@psudmant

@tobiasmarschal

@wwliao88

@cbioportal

@zevkronenberg

@pickettbd

@artofbiology

@HopsWolfram

@mjpchaisson

@DOEKBase
Check out SVbyEye: A new visualization tool to characterize structural variation among whole-genome assemblies (biorxiv.org/content/10.110…)

Detailed evaluation of remaining gaps in phased genome assemblies from human pangenome consortium (HPRC). We show that large repeat flanked inversions are among those the are the most difficult to assemble correctly. For more see: genome.cshlp.org/content/early/…
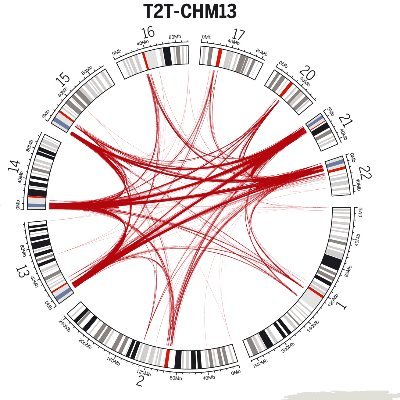
Here we prove that the complete human genome assembly (T2T-CHM13) is a better reference for inversion discovery than GRCh38. For instance, T2T reference is three times more likely to represent the correct orientation of the major human allele. See: genomebiology.biomedcentral.com/articles/10.11…
"Telomere-to-telomere assembly of diploid chromosomes with Verkko" now published 🎉 Great work @MikkoRautiaine3 @sergeynurk Brian Walenz @glennis_logsdon @PorubskyDavid @ArangRhie @EichlerLab @sergekoren! nature.com/articles/s4158…
The other on how much new we can learn about uoısɹǝʌuı polymorphism with a gapless human genome biorxiv.org/content/10.110…
We are excited to annouce bioRxiv release of two new manuscripts! One on critical evaluation of phased genome assemblies biorxiv.org/content/10.110… which offers insight into where state-of-the-art haplotype-resolved assemblies still struggle
To help unpack the diversity within SD sequence, we are excited about the future of technologies like: nature.com/articles/s4159…, github.com/marbl/verkko, and nature.com/articles/s4158… which bring the possibility of fully assembling human genomes within reach! #T2T #assembly
Finally, putting all of these elements together, we hope to tackle questions regarding human evolution and non-human primate assembly and lineage! nature.com/articles/s4158… and nature.com/articles/s4158… @yafmao92 @PorubskyDavid #nhp
In case you missed the interviews with Mitchell Vollger (@mrvollger), Glennis Logsdon (@glennis_logsdon) & Evan (@EichlerEE) on the #T2T papers, check it out! newsroom.uw.edu/news/some-hard… youtu.be/mPcovmMjFgc
Hello Twitter World! The Eichler Lab has decided to join the 21st century and get a Twitter account! We wanted to start this account out by tweeting about things we are excited about moving forward! #genomics #biology @HHMINEWS @uwgenome @EichlerEE
United States Trends
- 1. Mike 1,78 Mn posts
- 2. Serrano 237 B posts
- 3. #NetflixFight 70,6 B posts
- 4. Canelo 16 B posts
- 5. #netflixcrash 15,4 B posts
- 6. Father Time 10,7 B posts
- 7. Logan 77,3 B posts
- 8. Rosie Perez 14,7 B posts
- 9. He's 58 24 B posts
- 10. Shaq 15,8 B posts
- 11. Boxing 292 B posts
- 12. ROBBED 101 B posts
- 13. #buffering 10,8 B posts
- 14. My Netflix 82,2 B posts
- 15. Tori Kelly 5.130 posts
- 16. Roy Jones 7.093 posts
- 17. Ramos 70,2 B posts
- 18. Cedric 21,6 B posts
- 19. Gronk 6.560 posts
- 20. #cancelnetflix 4.915 posts
Who to follow
-
 Glennis Logsdon
Glennis Logsdon
@glennis_logsdon -
 EichlerLab
EichlerLab
@EichlerLab -
 Mitchell R. Vollger
Mitchell R. Vollger
@mrvollger -
 Stergachis Lab
Stergachis Lab
@stergachislab -
 Human Pangenome Reference Consortium
Human Pangenome Reference Consortium
@HumanPangenome -
 Peter Sudmant
Peter Sudmant
@psudmant -
 Tobias Marschall
Tobias Marschall
@tobiasmarschal -
 Wen-Wei Liao 廖玟崴
Wen-Wei Liao 廖玟崴
@wwliao88 -
 cBioPortal
cBioPortal
@cbioportal -
 Zev Kronenberg
Zev Kronenberg
@zevkronenberg -
 Brandon Pickett
Brandon Pickett
@pickettbd -
 Min Dai
Min Dai
@artofbiology -
 Wolfram Höps
Wolfram Höps
@HopsWolfram -
 Mark Chaisson
Mark Chaisson
@mjpchaisson -
 KBase
KBase
@DOEKBase
Something went wrong.
Something went wrong.