Chirag Jain
@chirgjainAssistant Professor @iiscbangalore. Previously @genome_gov, @GTcse and @IITDelhi
Similar User

@tobiasmarschal

@coxtonyj

@rrwick

@sergeynurk

@sedlazeck

@BenedictPaten

@MishaKolmogorov

@kishwarshafin

@pashadag

@RayanChikhi

@BQPMalfoy

@veli_makinen

@r_sabarinathan

@aquaskyline

@IliaMinkin
Happy to see our method for T2T genome assembly published! It addresses an important limitation of string graph, that is, the contained reads. Led by @skamath5e "Telomere-to-telomere assembly by preserving contained reads" doi.org/10.1101/gr.279…
SPECIAL ISSUE! This month @genomeresearch publishes a diverse collection of research and review articles in a special issue highlighting advances in long-read sequencing applications in biology and medicine. tinyurl.com/Genome-Res-34-….

🌟 3 Days to Go! 🌟 Last chance to apply for #ACMIndia Winter Schools 2024! Dive into advanced tech with top experts from academia & industry! 🚀 Limited seats – 40 per school! 🔗 Apply now: lnkd.in/dHDekPA2 👉 Apply now and share with friends!

Pangene now published in Bioinformatics: doi.org/10.1093/bioinf…. In addition to showcasing applications (see the 17q21.31 inversion below), we also reviewed the theoretical formulation of bidirected graphs and discussed the definition and the finding of "bubbles" in such graphs.

Preprint on Exploring gene content with pangenome gene graphs: arxiv.org/abs/2402.16185. It describes pangene for building gene graphs and for calling gene-level variations which can be found at pangene.bioinweb.org. Pleasant collaboration with @maxgmarin and @MahaFarhat
Our recent work "Haplotype-aware sequence alignment to pangenome graphs" is now online at @genomeresearch (RECOMB'24 issue). It introduces new algorithms and complexity results for the problem. Led by @GhanshyamChand5 doi.org/10.1101/gr.279…

We send our appreciation to the speakers, poster presenters, participants, student and staff volunteers who contributed to the resounding success of the BDBio 2024 held last week! @iiscbangalore @cdsiisc




Our review paper on "Genome assembly in the telomere-to-telomere era" published online in @NatureRevGenet It amazes me how much assembly has progressed in the past four years – often ~100X improvement in contiguity and base accuracy and with haplotypes and hard regions resolved!
Genome assembly in the telomere-to-telomere era go.nature.com/4aLqIED #Review by Heng Li @lh3lh3 & Richard Durbin @richard_durbin @DanaFarber @HarvardDBMI @Cambridge_Uni
Symposium on Big Data Algorithms for Biology (May 31-June 1 2024) is now open for registration and abstract submission [Deadline: Apr 20] bdbio.in Join us for the engaging talks, poster presentations, and panel discussions in various topics of comp. biology

Gapless assembly of complete human and plant chromosomes using only nanopore sequencing | bioRxivc Happy @keygene could contribute to this work. @nanopore #T2T #duolex biorxiv.org/content/10.110…
The CDS department at IISC Bangalore invites applications for its summer internship programme, aimed at students pursuing the relevant branch of engineering! The deadline for the application is 15th March 2024. cds.iisc.ac.in/call-for-cds-s… @cdsiisc #cdsinternship2024

Join us at the second annual Symposium on Big Data Algorithms for Biology (BDBio) on May 31-June 1, 2024 @iiscbangalore The 2-day event will feature cutting-edge research in computational biology. Registration opens on March 20. More details here: bdbio.in

Happy to present an initial draft of a telomere-to-telomere diploid Indian genome. A joint effort of Jianjun Liu's and my lab spearheaded by @msprasad693 and @JosipaLipovac github.com/lbcb-sci/I002C or github.com/LHG-GG/I002C
Are nanopore UL reads only long reads we need? We developed Herro github.com/lbcb-sci/herro AI error correction model that can correct reads to accuracy above Q30 while trying to keep informative positions intact. w/ @domstanojevic @DehuiLin @sergeynurk @PaolaFlorezdeS @nanopore
The list of #RECOMB2024 Accepted Papers is now live: recomb.org/recomb2024/acc…. A huge congrats to all the authors! @RECOMBconf

Happy to see that the RAFT method was useful here @skamath5e github.com/at-cg/RAFT
SM: @nanopore only T2T assembly enable by highly accurate ultra-long reads using off the shelves tools #NanoporeConf

RECOMB-Seq 2024 Call for Papers is out! recomb-seq.github.io/papers/ Submit via EasyChair. Select “RECOMBSeq 2024” track within RECOMB conference at easychair.org/my/conference?… Calendar: Abstract registration: January 30, 2024 Final submission: February 06, 2024 Event: April 27-28, 2024
What’s more, telomere-to-telomere (#t2t) assemblies now achievable with JUST simplex. Q28 simplex data is accurate enough. You do not need data from any other platform — paving the way for @nanopore T2T assembly, using just simplex data. #nanoporeconf 1/2

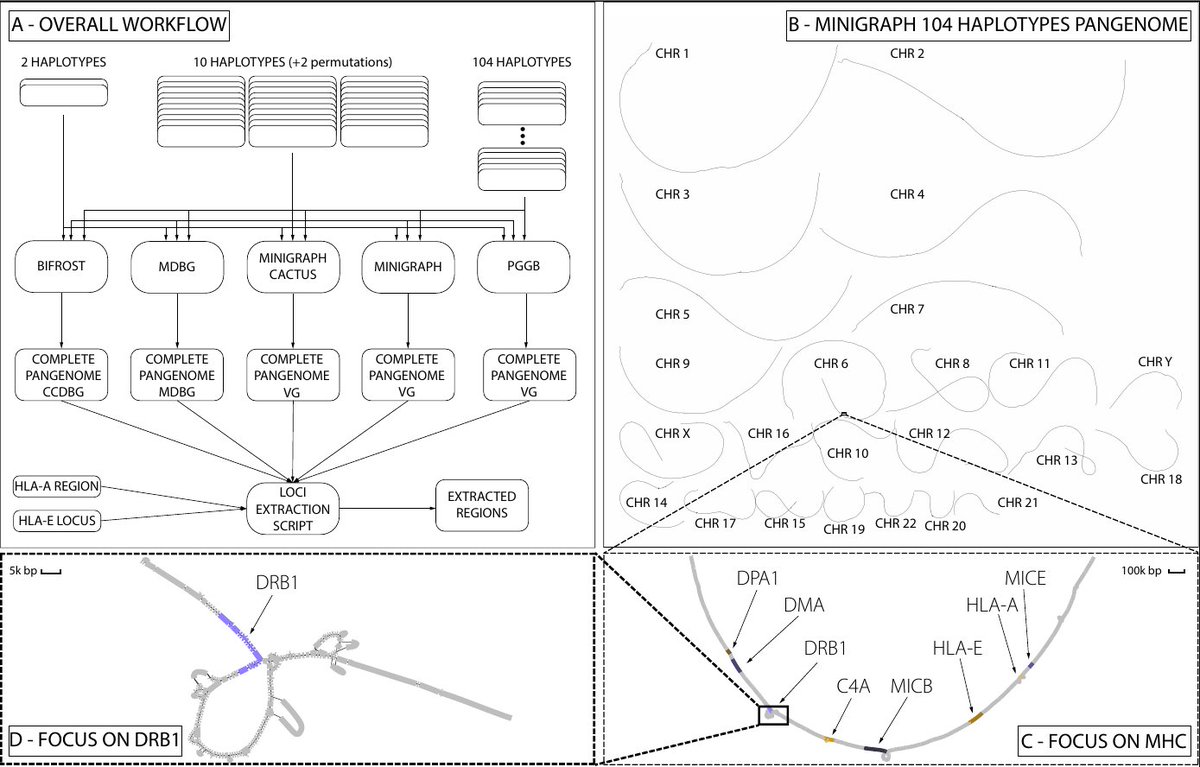
Comparing graph construction methods for complete human pangenomes, by @frnandreac and @yoann_dufresne genomebiology.biomedcentral.com/articles/10.11… With a focus on subgraph extraction of loci of interest. Preprint thread:
The construction of large cohorts human pangenome graphs is a challenging task. We selected 5 different state-of-the art tools that generate variation graphs or De Bruijn graphs and used them to construct a human pangenome of 104 high-quality human haplotype assemblies. (1/4)
Recent work led by @GhanshyamChand5 introduces new formulations and algorithms for approximate pattern matching to haplotype-resolved pangenome references biorxiv.org/content/10.110…
The latest work from our lab, led by Sudhanva Kamath and Mehak Bindra, addresses the challenge of "contained reads" in long-read assembly. The preprint is now up: biorxiv.org/content/10.110…

@genomeresearch is now encouraging submissions for a Special Issue on Long-read DNA Sequencing Applications in Biology and Medicine, Guest Edited by @anaconesa, @ahoischen, and @sedlazeck bit.ly/grcallforpapers #longreads #T2T #LongTREC #Hitseq #ASHG23

United States Trends
- 1. Pam Bondi 147 B posts
- 2. Jameis Winston 6.240 posts
- 3. Browns 35,6 B posts
- 4. #911onABC 22,5 B posts
- 5. Gaetz 936 B posts
- 6. Brian Kelly 3.655 posts
- 7. Eddie 44,3 B posts
- 8. #PITvsCLE 3.140 posts
- 9. #TNFonPrime 2.134 posts
- 10. Bryce Underwood 20 B posts
- 11. Fields 45,7 B posts
- 12. #GoBlue 6.410 posts
- 13. Pickens 4.881 posts
- 14. Brad 34,7 B posts
- 15. #DawgPound 2.947 posts
- 16. Michigan 70,2 B posts
- 17. Lamelo 6.363 posts
- 18. Al Michaels N/A
- 19. Nick Chubb 1.424 posts
- 20. Boswell 2.024 posts
Who to follow
-
 Tobias Marschall
Tobias Marschall
@tobiasmarschal -
 Tony Cox coxtonyj @genomic.social .bsky.social
Tony Cox coxtonyj @genomic.social .bsky.social
@coxtonyj -
 Ryan Wick
Ryan Wick
@rrwick -
 Sergey Nurk 🇺🇦
Sergey Nurk 🇺🇦
@sergeynurk -
 Fritz Sedlazeck
Fritz Sedlazeck
@sedlazeck -
 Benedict Paten
Benedict Paten
@BenedictPaten -
 Misha Kolmogorov
Misha Kolmogorov
@MishaKolmogorov -
 Kishwar
Kishwar
@kishwarshafin -
 PM @[email protected] @pashadag.bsky.social
PM @[email protected] @pashadag.bsky.social
@pashadag -
 Rayan Chikhi
Rayan Chikhi
@RayanChikhi -
 Malfoy
Malfoy
@BQPMalfoy -
 Veli Mäkinen
Veli Mäkinen
@veli_makinen -
 Sabarinathan Radhakrishnan
Sabarinathan Radhakrishnan
@r_sabarinathan -
 Ruibang Laurent Luo
Ruibang Laurent Luo
@aquaskyline -
 Ilia Minkin
Ilia Minkin
@IliaMinkin
Something went wrong.
Something went wrong.