Fabian
@FabianJetzingerPhD student in Bioinformatics @ BioBam, studying long-read transcriptomics; he/him
Welcoming visitors Yalan and Fran, from @LongtrecEU @MSCActions project, with a fun Journal Club discussing Splam, a deep-learning-based splice site predictor #lrRNAseq genomebiology.biomedcentral.com/articles/10.11…

Wrapping up the @SEBiBC_es congress, very proud of the lab's contributions! If you didn't get to see them, check out @Tian_Yuan_Liu's, @CarolMonzo's, Quique's and Alejandro's posters below 🫶 @i2sysbio @BCBHubCSIC

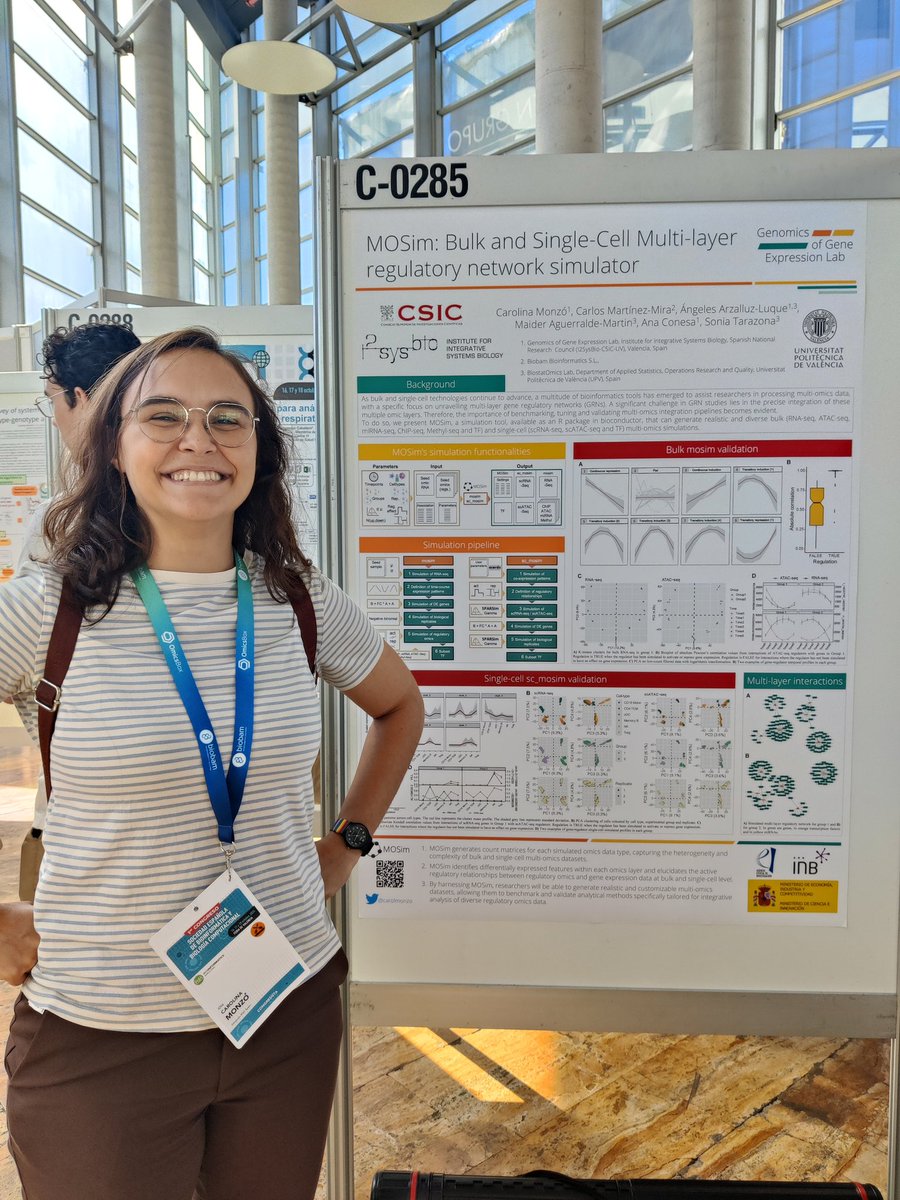


Very proud of @Conesa_Lab postdoc @CarolMonzo for not only getting selected as a finalist for best PhD thesis award (done at @MPIAGE) but also winning the best poster award from the @SEBiBC_es for our latest software! @i2sysbio @BCBHubCSIC Awesome job! 🏄♀️


Giant thank you to @SEBiBC_es for this great award! And to the amazing team of people working on MOSim for their indispensable input 🤗🎉
Por último y no menos importante premio al mejor póster para @CarolMonzo por el desarrollo de MOSIM. Enhorabuena!!!

It was an honor to be recognized by the Spanish Society for Bioinformatics and Computational Biology as a finalist for the Oswaldo Trelles award to the Best PhD thesis! Big thanks to the @MPIAGE, Linda Partridge, @AndreasBeyer012 and the LP PhD students for all their support!
El premio Oswaldo Trelles ha dejado también una mención especial a la tesis de @CarolMonzo que ha recibido un diploma durante la ceremonia. Enhorabuena!!

Excited to have some of my work contribute to Ana's great talk! It's been great to see so much interest in our work! Also excited about the #longtrec Long-Read Transcriptomics workshop tomorrow afternoon! #LRUA24
Coolest talk starting!!! @anaconesa will tell us about long reads transcriptomics, biases, experimental design and the SQANTI-verse!!! @BCBHubCSIC @i2sysbio @CSIC #LRUA24


Now is the time to submit abstracts for #LRUA24! This is a great opportunity to share your long-read work with leading experts. Both talks and posters will be selected For submission, visit the conference website: lrua2024.se Abstract deadline is Aug 21st

Reading the reviews of a rejected proposal makes my blood boil. R3 states that the project doesn't develop a new methodology but "only" novel bioinformatics methods. How long will this disregard for computational research continue? Try to do your sequencing data analysis by hand!
First day of my 🇸🇪 secondment with @krsahlin at the @scilifelab! Already had productive discussions, ideas for longer-term collaborations, and met the princess of Sweden at lunch (kind of). Super grateful for @LongtrecEU for providing such amazing opportunities to learn!

Finally, I love projects like this, where researchers love their animals and it shows in the quality and dignity of their work! Would be nice if people working on human data loved the organisms they are working on the same way… x.com/FZanarello/sta… 25/25
Bioinformatics has never been so wild. Sampling Xatardia plant to build and annotate its genome using long read sequencing. Supervised by @Silvia_Torello and @RodericGuigo , guided by @pladevallclara and Manel. @CRGenomica @GuigoLab, @LongtrecEU @Andorra_RI


Amazing work by @TheBrooksLab, @Anaconesa lab, and the multitude of their students and students in other labs who led the LRGASP project that pushed forward long-read technology and analysis benchmarking! x.com/anaconesa/stat… 24/
I´m thrilled to see our #LRGASP paper published in @naturemethods today. This is the work of many to benchmark long-read methods for transcriptomics, and a must-read paper for those using lrRNA-seq. Special thnks to @FJPardoPalacios and @TheBrooksLab (1/6) rdcu.be/dJ9ev

SQANTI3 release alert🚨 Are you a SQANTI3 user? Go ahead and update your installation to v5.2.2, our newest release includes bug fixes, updates and improvements in classification of monoexons, genic introns, NIC and genic categories. Don't miss the update! github.com/ConesaLab/SQAN…
LongTREC students discussing with the coordinator @anaconesa to plan the Long-read transcriptomics workshop for the #LRUA24 in Uppsala Don't miss this event lrua2024.se/program/

Last chance to sign up to tomorrow's webinar on End-to-End Long-Read RNA-Seq Analysis in OmicsBox! There are two sessions (starting at 9:30 and 18:30 CEST), hope to see many of you there!
Lasts days to join our Long-Reads RNA-Seq Analysis webinar! #longreads #transcriptomics #bioinformatics #genomics events.zoom.us/eo/AvwSOjeTUdU…
I´m thrilled to see our #LRGASP paper published in @naturemethods today. This is the work of many to benchmark long-read methods for transcriptomics, and a must-read paper for those using lrRNA-seq. Special thnks to @FJPardoPalacios and @TheBrooksLab (1/6) rdcu.be/dJ9ev

LongTREC members attended London Calling 2024 #nanoporeconf last week to meet a very excited community interested in the newest developments of long-read technologies! @AinoJarvelin @nanopore @FabianJetzinger @bio_bam @logan_mulroney @IITalk Satrio Benowo @UniofNottingham

Some incredible stories and words of wisdom to the next generation of scientists from Dave, Dan, Mark, and Hagan. Having the space to be curious and chase strange observations is invaluable, and when science is the most fun! #nanoporeconf

There is wine, music, and face painting at the #nanoporeconf evening program... Need i say more? @nanopore @bio_bam

Great first day, participating in a workshop on EPI2ME at @nanopore London Calling 2024, meeting fascinating people and learning lots, looking forward to tomorrow! Thanks for making it possible @LongtrecEU & @bio_bam! #nanoporeconf #longtrec
United States Trends
- 1. Gaetz 771 B posts
- 2. Ken Paxton 10,2 B posts
- 3. DeSantis 23,4 B posts
- 4. Attorney General 203 B posts
- 5. Volvo 15,8 B posts
- 6. Gary Gensler 28,1 B posts
- 7. Mike Davis 3.001 posts
- 8. 119th Congress 6.267 posts
- 9. Andrew Bailey 2.689 posts
- 10. Mark Levin 1.191 posts
- 11. Rubio's Senate 8.937 posts
- 12. ICBM 224 B posts
- 13. Dragon Believer 1.467 posts
- 14. Denver 41,7 B posts
- 15. The ICC 385 B posts
- 16. Jussie Smollett 19,4 B posts
- 17. Netanyahu 756 B posts
- 18. $SOLCAT 5.418 posts
- 19. Flat 56 B posts
- 20. Murkowski 23,3 B posts
Something went wrong.
Something went wrong.