Luis M Rodriguez-R
@_lrr_Feminist dancer who happens to be into microbial ecology. Asst. Prof. at @uniinnsbruck. Cat human. Developer of @MicroGenomes MiGA. He/him. https://t.co/lv7OcGddjC
Similar User

@MattagenOlmics

@BanfieldLab

@CMR_QUT

@PhilHugenholtz

@Thuls_M

@OrtnerNadine

@merenbey

@ChristianRinke

@hei_anni

@alejandrorgijon

@sdbn

@luisarchaeota

@campanarostef

@ViorelaDan

@quendi
New species of #archaea from arthropods described under the #SeqCode have been finally approved! Since genomes were used as type material for species description, many taxa previously known only from metagenomics now have official species protologues. doi.org/10.57973/seqco…

This seems to be a true evidence that intraspecifically bacterial species are organized as discrete genomic groups. Genomovars exist though! @_lrr_ @TomeuViver @CotoOrellana
A genomic sequence discontinuity within species has been detected across Bacteria around 99.5% ANI. @_lrr_ and coauthors propose its use for the definition of genomovars and compare them to sequence types and other infra-specific groups. @CotoOrellana journals.asm.org/doi/10.1128/mb…

A genomic sequence discontinuity within species has been detected across Bacteria around 99.5% ANI. @_lrr_ and coauthors propose its use for the definition of genomovars and compare them to sequence types and other infra-specific groups. @CotoOrellana journals.asm.org/doi/10.1128/mb…

@seq_code is back again on #BISMiSLive with @_lrr_ delivering a talk on the #SeqCode registry. Register at us02web.zoom.us/webinar/regist… @ICSP_news @fanus_venter @PhilHugenholtz @Leibniz_DSMZ_en @m_chuvochina @mjpallen @BccmCollections @ISME_microbes

I'm excited to be selected for HPC Achievement Award by @NERSC, for the FastANI project doi.org/10.1038/s41467… Thanks to the project team @_lrr_, @aphillippy, Srinivas Aluru, Kostas Konstantinidis, and @NERSC for enabling this.
Congratulations to the 2022 @NERSC High Performance Computing Achievement Award winners! These awards honor early-career researchers who've made significant contributions to scientific computation using NERSC resources: bit.ly/NERSCAchieveme… @BerkeleyLab #HPC

Von sozialen Robotern bis zu personalisierter Medizin – Universität Innsbruck uibk.ac.at/de/newsroom/20…
65.000 neue Namen per Mausklick: #Algorithmen könnten die Benennung von Mikroben erheblich beschleunigen. Das zeigen zwei neue Publikationen mit Beteiligung von Miguel Rodríguez-Rojas @_lrr_ vom Institut für #Mikrobiologie und @DiSC_uibk: uibk.ac.at/de/newsroom/20… #ibktwit #SeqCode
🧬 Durch verbesserte DNA-Sequenzierung wurden so viele Prokaryoten entdeckt, dass man mit dem Benennen kaum nachkommt und viele Mikroben einen Code erhalten. Neue Algorithmen könnten helfen @_lrr_ @uniinnsbruck 🔗doi.org/10.1038/s41564… 🔗doi.org/10.1099/ijsem.… © Fabian Oswald

Luis Miguel Rodríguez-Rojas (@_lrr_) von der @uniinnsbruck war an zwei Publikationen beteiligt, welche die Namensgebung von Mikroorganimen erheblich beschleunigen könnten. #SeqCode idwf.de/-DEAuAC
ICYMI: SeqCode: a nomenclatural code for prokaryotes described from sequence data. By @AlexJProbst @marike_palmer @_lrr_; @seq_code @MurrayLab1 @donovan_parks @m_chuvochina @ramonibus @PhilHugenholtz @alr_thermophile @fanus_venter & colleagues. nature.com/articles/s4156…
Need a name for your new (prokaryotic) find? 🦠 Explore the @Seq_Code initiative with @NatureMicrobiol nature.com/articles/s4156…
A paper published in #IJSEM proposes a new method of naming newly discovered microbe species, using a computer programme to generate tens of thousands of names. Learn more: microb.io/3xDQ1aa
Over 65,000 unnamed Archaea and Bacteria named using well-formed Latin names generated by scripts in this publication. Interesting article by @mjpallen, @_lrr_, and @happy_khan doi.org/10.1099/ijsem.…
Have you heard of SeqCode? If not, may we introduce you 👉SeqCode: a nomenclatural code for prokaryotes described from sequence data @seq_code nature.com/articles/s4156…
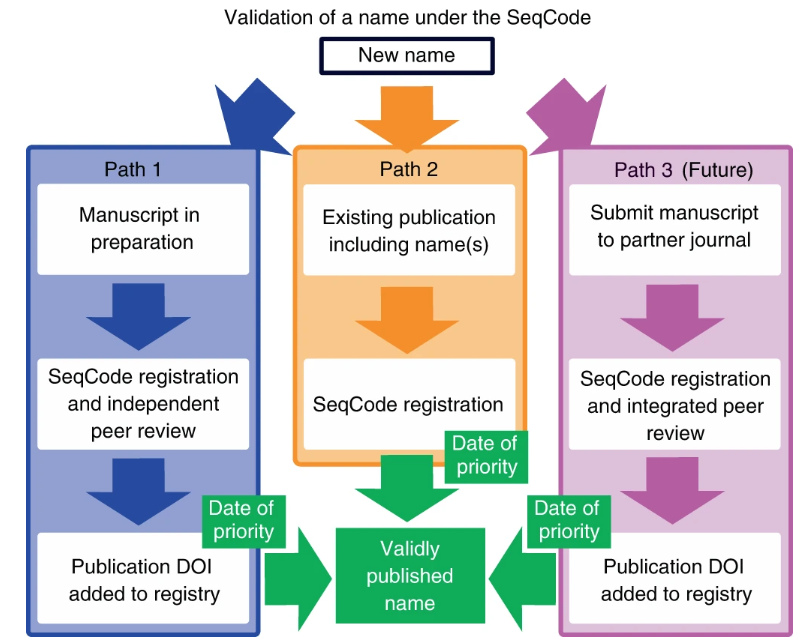
Dear all, we are happy to announce that the official publication of the #SeqCode is now live. Please circulate this paper among your colleagues, and let's open together a new chapter in the prokaryotic nomenclature... nature.com/articles/s4156…
¿Te interesa conocer qué sucede con los microorganismos después de un derrame de petróleo? Entonces no te pierdas la charla del profesor Luis Miguel Rodríguez @_lrr_ @uniinnsbruck Transmisión en vivo por nuestro canal de YouTube👉🏻 youtu.be/9btAQp2psz4

United States Trends
- 1. $RWA 15,5 B posts
- 2. $MXNBC 1.459 posts
- 3. Saquon 98,9 B posts
- 4. #MondayVibes 3.726 posts
- 5. Good Monday 29,9 B posts
- 6. #DatGirlisIZNA 6.735 posts
- 7. #이즈나_음원발매_데뷔 6.659 posts
- 8. IZNA DEBUT OUT NOW 6.612 posts
- 9. Eagles 116 B posts
- 10. Idleness 1.928 posts
- 11. Rams 41,6 B posts
- 12. Snoop 12,1 B posts
- 13. #WayV_FREQUENCY 38,9 B posts
- 14. Steve Lacy 3.249 posts
- 15. Chris Chan 10,7 B posts
- 16. #BaddiesMidwest 15,6 B posts
- 17. Brandon Graham 11,8 B posts
- 18. Uruguay 185 B posts
- 19. Taiga 5.034 posts
- 20. Jela 4.408 posts
Who to follow
-
 Matt Olm
Matt Olm
@MattagenOlmics -
 The Banfield Lab
The Banfield Lab
@BanfieldLab -
 Centre for Microbiome Research
Centre for Microbiome Research
@CMR_QUT -
 Phil Hugenholtz
Phil Hugenholtz
@PhilHugenholtz -
 Thulani Makhalanyane
Thulani Makhalanyane
@Thuls_M -
 Nadine Ortner
Nadine Ortner
@OrtnerNadine -
 A. Murat Eren (Meren)
A. Murat Eren (Meren)
@merenbey -
 Chris Rinke
Chris Rinke
@ChristianRinke -
 Anna
Anna
@hei_anni -
 Alejandro Rodríguez-Gijón
Alejandro Rodríguez-Gijón
@alejandrorgijon -
 San Diego Biotech Networks
San Diego Biotech Networks
@sdbn -
 Luis E. Valentín-Alvarado
Luis E. Valentín-Alvarado
@luisarchaeota -
 stefano campanaro
stefano campanaro
@campanarostef -
 Viorela Dan
Viorela Dan
@ViorelaDan -
 Trina McMahon 🌪
Trina McMahon 🌪
@quendi
Something went wrong.
Something went wrong.