
Feng Xiao
@FengXiao1000Postdoc@Boston Children's Hospital, Harvard Medical School; Stem cell biologist; Dissecting genetic variants in heart disease with iPSC model and CRISPR.
Similar User

@Transactivator

@TheSurajKannan

@leo_jwu

@MingxiaGu

@SimonMiao6

@robin_canac

@chun_liu_86

@njvdn

@oscarabilez

@Deqiang8

@twinsmc18

@Thomaswei737

@arianneinthelab

@CacaceLucena

@JihyunJang_JJ
Excited to share that my first postdoctoral work is now online @NatureGenet: rdcu.be/dy7i9. We functionally dissect human cardiac enhancers and address how noncoding variants contribute to congenital heart disease. Thanks to all the people who contribute to this work.

In the latest issue! Saturation mutagenesis-reinforced functional assays for disease-related genes dlvr.it/TGCWq8
Multimodal scanning of genetic variants with base and prime editing go.nature.com/3O6wQ0O

VISTA Enhancer browser: an updated database of tissue-specific developmental enhancers. #DevelopmentalEnhancers @NAR_Open academic.oup.com/nar/advance-ar…
Our new paper on epigenome editing is out! We mapped which effector domains regulate transcription across genomic, cell type and DNA-binding domain contexts. Then we built new gene repressor and activator tools with improved efficiency & deliverability nature.com/articles/s4158…
I’m thrilled to share that our paper “Machine-guided design of cell-type-targeting cis-regulatory elements” was published online in @Nature nature.com/articles/s4158… #genomics #MachineLearning #SyntheticBiology #bioengineering 1/9
SUPER excited to share that my latest research has been published in @CellGenomics! Check out the thread below to explore our findings on the distinct gene regulatory programs that drive human cardiomyocyte development (1/n) 🫀 cell.com/cell-genomics/…

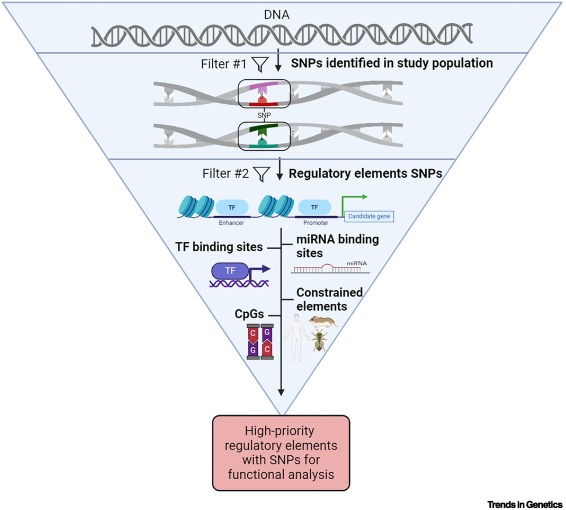
A bioinformatics toolbox to prioritize causal genetic variants in candidate regions dlvr.it/TFNTNS
In the latest issue! High-resolution functional mapping of RAD51C by saturation genome editing dlvr.it/TF2KRD
Genomic tech dev is my favorite area of science, and today our paper describing our multiplex CRISPRa screening method to identify cell type specific regulatory elements is published! rdcu.be/dUnoq
Read about our development of antisense oligo therapy for calmodulinopathy, an inherited arrhythmia, in the Boston Children's Hospital Answers blog: answers.childrenshospital.org/genetic-treatm… manuscript: ahajournals.org/doi/abs/10.116…

Read my latest #research on #GeneTherapy for #HeartFailure, published in @natBME A miniaturized version of the dyad-stabilizing protein CMYA5 mitigates heart failure in a mouse model. Read here: rdcu.be/dS3XD
Recordings from the Mutational Scanning Symposium are now available! #VariantEffect24 View them on our Youtube channel : youtube.com/@cmap_cegs8443…

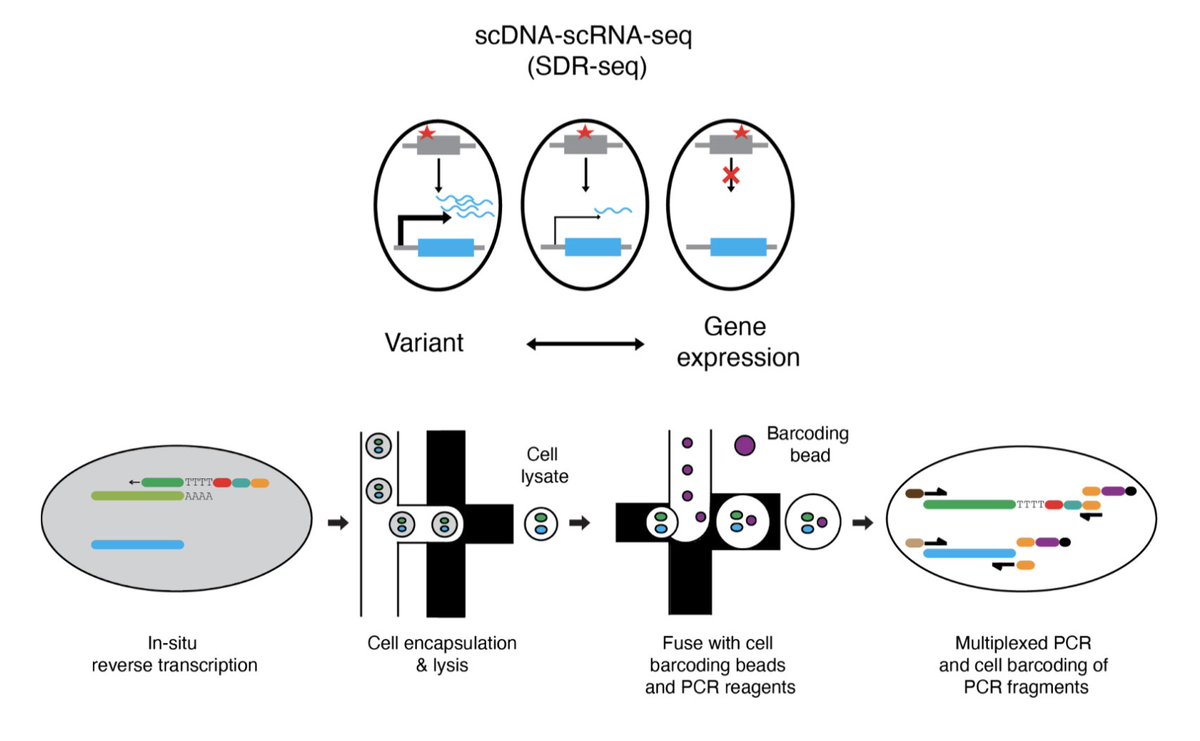
We are very happy to share our preprint (D. Lindenhofer et al.) of a high-throughput targeted scDNA-scRNA-seq (SDR-seq) method. Impactful for any application with a need to associate coding and non-coding variants to gene expression in tissue samples and cell culture systems.
Characterizing >100,000 sequences in primary neurons and organoids and deep learning to decode regulatory activity. Amazing work from @Chengyu_Deng_, @seawhalen, Katie Pollard, @LabNowakowski and others as part of #PsychENCODE in @ScienceMagazine science.org/doi/10.1126/sc…
We've found that the cell adherens junction is crucial for cardiomyocyte development through the NF2-YAP-AMOT protein complex @NUSMedicine @NUSResearch @CvdNus @TheFooLab @Meekelee
GoT-ChA! I am thrilled to announce that our paper "Mapping Genotypes to Chromatin Accessibility Profiles in Single Cells" is now out in @Nature !! 👩🔬👨🔬🧬If you want to link somatic mutations to epigenetic changes directly in patient samples: nature.com/articles/s4158… 👇🧵
Nice correlation between massively parallel reporter assays (MPRAs) and mouse enhancer assays and great neurodev enhancer catalog. Great work and collaboration from @michaelkosicki, @DianneLaboy, @LenPennacchio and many others. biorxiv.org/content/10.110…
Tissue-specific enhancer–gene maps from multimodal single-cell data identify causal disease alleles nature.com/articles/s4158… #journals #feedly
Ever considered measuring genetic variant effects with prime editing but hesitated due to low and variable editing? Last month, 3 papers showed that large-scale screens can work 💪. A 🧵 comparing the papers of the @fjsanchezrivera lab, the @bsadamson lab, and @TheGenomeLab

Join the Novo Nordisk Foundation Center for Genomic Mechanisms of Disease at Broad Institute on Tuesday, March 19 for the Variant to Function Symposium. Attend in-person at Broad, or virtually. More info: broad.io/v2f

United States Trends
- 1. #UFC309 88,4 B posts
- 2. Tatum 18,1 B posts
- 3. #MissUniverse 298 B posts
- 4. Oregon 29,4 B posts
- 5. Beck 16,8 B posts
- 6. Dan Lanning N/A
- 7. Dinamarca 18,4 B posts
- 8. Wisconsin 47,2 B posts
- 9. gracie 15,7 B posts
- 10. Locke 4.835 posts
- 11. Nigeria 218 B posts
- 12. Llontop 14,1 B posts
- 13. Ruffy 17,2 B posts
- 14. Brian Kelly 10,3 B posts
- 15. Mac Miller N/A
- 16. Jim Miller 10,4 B posts
- 17. Inside the NBA 6.668 posts
- 18. Tailandia 11,2 B posts
- 19. Dalton Knecht 6.056 posts
- 20. #AEWCollision 14 B posts
Who to follow
-
 Eric N. Olson
Eric N. Olson
@Transactivator -
 Suraj Kannan
Suraj Kannan
@TheSurajKannan -
 Jun Wu Lab
Jun Wu Lab
@leo_jwu -
 Mingxia Gu
Mingxia Gu
@MingxiaGu -
 Simon Miao
Simon Miao
@SimonMiao6 -
 Robin Canac
Robin Canac
@robin_canac -
 Chun
Chun
@chun_liu_86 -
 Nathan VanDusen
Nathan VanDusen
@njvdn -
 Oscar J. Abilez
Oscar J. Abilez
@oscarabilez -
 Deqiang Li(DQ)
Deqiang Li(DQ)
@Deqiang8 -
 Simon Shen
Simon Shen
@twinsmc18 -
 Thomas Wei
Thomas Wei
@Thomaswei737 -
 Arianne Caudal
Arianne Caudal
@arianneinthelab -
 Antonio Lucena-Cacace
Antonio Lucena-Cacace
@CacaceLucena -
 Jihyun Jang
Jihyun Jang
@JihyunJang_JJ
Something went wrong.
Something went wrong.













































































































