Similar User

@sydshaffer

@cojiberries

@QinZhu_1

@hao_wu_7

@ericjoycelab

@SRouhanifard

@CreminsLab

@yogeshgoyallab

@jisaacmurray

@ophirshalem

@dsjpark

@BethBGenetics

@hsiung_chris

@mrlawrencepaul

@lisanwang
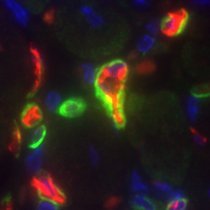
What if you could find out the history of signaling activity in each cell with a single round of imaging? Wouldn't that be incredible? Well, we thought so too. And that's what we set out to achieve with INSCRIBE in our new preprint. Let’s dive in! 🚀🔬 biorxiv.org/content/10.110…
After precisely 4 years of grad school, I'm excited to share that the FIRST PEER REVIEWED PAPER FROM MY PHD HAS LANDED! cell.com/cell-systems/f… Indulge me in some back-story. 🧵
Transcriptional adaptation is so cool! and now we know how widespread it is- check out the paper for more. Very grateful to have worked with and learned so much from @ian_mellis !
Thrilled to share our latest @GenomeBiology led by @ian_mellis identifying widespread presence of transcriptional adaptation *really cool biology* in mammals w/ data mining & stochastic modeling thx @NU_CDB @NUSynBio @czbiohub @NITMB_Chicago @LurieCancer genomebiology.biomedcentral.com/articles/10.11…
Thrilled to share our latest @GenomeBiology led by @ian_mellis identifying widespread presence of transcriptional adaptation *really cool biology* in mammals w/ data mining & stochastic modeling thx @NU_CDB @NUSynBio @czbiohub @NITMB_Chicago @LurieCancer genomebiology.biomedcentral.com/articles/10.11…
My PhD work on harnessing the brain parasite Toxoplasma gondii for protein delivery to the brain is out in @NatureMicrobiol ! nature.com/articles/s4156…
Excited to share our new @biorxivpreprint in which we demonstrate the formation and maintenance of cellular memories in melanoma during acquisition of therapy resistance - dependent on the transcription factor AP-1 biorxiv.org/content/10.110… (1/9)
Interested in knowing how well doublet detection algorithms do and extract real singlets from scRNA-seq data? excited to share first research paper from the Goyal lab, a Herculean effort by @madelinemelzer & Charles in @CellGenomics! @NUFeinbergMed @NUSynBio @NU_CDB @czbiohub
Interested in reliably and robustly extracting ground-truth singlets in your scRNA-seq datasets? Excited to share a project from @yogeshgoyallab and Ziyang Zhang, using DNA barcodes to identify doublets in single-cell RNA sequencing data cell.com/cell-genomics/… (1/

Interested in reliably and robustly extracting ground-truth singlets in your scRNA-seq datasets? Excited to share a project from @yogeshgoyallab and Ziyang Zhang, using DNA barcodes to identify doublets in single-cell RNA sequencing data cell.com/cell-genomics/… (1/

Truly amazing PI, very talented and collegial lab members, exciting and diverse scientific ideas--if I were looking for a new position in the fields he mentions, I would definitely write to @yogeshgoyallab!
Interested in single-cell fate tracing in a variety of cancer and stem cell contexts? We have postdoc positions funded by Pew, CZI, and others. Reach out if interested!
Sharing the lab's first preprint! This was a herculean team effort led by Abby, Roshan and Jessie in the lab with contributions from additional past members. We describe a molecular recorder for tracing signaling histories in cells during development biorxiv.org/content/10.110…
Our latest story on antigen presentation during #zebrafish #heart #regeneration is finally out! @StainierLab nature.com/articles/s4146…
Transcriptional responses of genes to master regulators can take complex non-linear forms. We wouldn't have been able to show this without systematically modulating TF dosage to a range of subtle to strong effects. So happy finally to see part of my postdoc work out! Great 🧵👇
We know the importance of subtle gene dosage variation, and yet the main tool of molbio is gene KO/KD. In our new preprint, @JuliaDiumenge introduced subtle modulations of transcription factor dosage by CRISPRi/a, and quantified responses by scRNA-seq. 1/ biorxiv.org/content/10.110…
A dream of our lab has been to image the full central dogma from a single endogenous gene, all live and with single molecule resolution. After many years we are happy to unveil a beautiful cell line that makes it possible. Check out our preprint (biorxiv.org/content/10.110…)! (1/n)
United States Trends
- 1. Colts 14,4 B posts
- 2. Brian Robinson 2.085 posts
- 3. #ai16z 2.642 posts
- 4. Aaron Jones 1.627 posts
- 5. Jonnu Smith 1.035 posts
- 6. #RaiseHail 4.216 posts
- 7. Keenan Allen 1.628 posts
- 8. Amorim 135 B posts
- 9. Brandon Aubrey N/A
- 10. Caleb Williams 3.710 posts
- 11. #SKOL 2.940 posts
- 12. Sean Tucker N/A
- 13. Noah Gray 1.132 posts
- 14. Bryce Young 1.595 posts
- 15. Nick Westbrook N/A
- 16. Geraldo 9.861 posts
- 17. Tolbert N/A
- 18. Rashford 39 B posts
- 19. #TitanUp N/A
- 20. #DALvsWAS 1.563 posts
Who to follow
-
 Sydney Shaffer
Sydney Shaffer
@sydshaffer -
 Connie Jiang
Connie Jiang
@cojiberries -
 Qin Zhu
Qin Zhu
@QinZhu_1 -
 Hao Wu | 吴昊
Hao Wu | 吴昊
@hao_wu_7 -
 Eric Joyce
Eric Joyce
@ericjoycelab -
 Sara Rouhanifard
Sara Rouhanifard
@SRouhanifard -
 Jenn Cremins
Jenn Cremins
@CreminsLab -
 Yogesh Goyal
Yogesh Goyal
@yogeshgoyallab -
 John Murray
John Murray
@jisaacmurray -
 Ophir Shalem
Ophir Shalem
@ophirshalem -
 Daniel S. Park
Daniel S. Park
@dsjpark -
 Elizabeth (Beth) Burton
Elizabeth (Beth) Burton
@BethBGenetics -
 Chris Hsiung 熊
Chris Hsiung 熊
@hsiung_chris -
 Matthew R. Lawrence-Paul
Matthew R. Lawrence-Paul
@mrlawrencepaul -
 Li-San Wang
Li-San Wang
@lisanwang
Something went wrong.
Something went wrong.