Devon Ryan
@dpryan79An American in Germany, an ion channel guy in bioinformatics, a proponent of scientific skepticism in an irrational world. @[email protected]
Similar User

@johanneskoester

@nf_core

@biorxiv_bioinfo

@torstenseemann

@StevenSalzberg1

@OliverStegle

@erikgarrison

@nomad421

@AliciaOshlack

@markrobinsonca

@mike_schatz

@wolfgangkhuber

@brent_p

@PaoloDiTommaso

@RayanChikhi
Alevin-fry-atac enables rapid and memory frugal mapping of single-cell ATAC-seq data using virtual colors for accurate genomic ... biorxiv.org/cgi/content/sh… #biorxiv_bioinfo
Glad they learned their lesson there. Did anyone like 0/0 for no calls?
GATK 4.6.0.0 reverts back to ./. for no-calls instead of 0/0 with DP=0 github.com/broadinstitute…
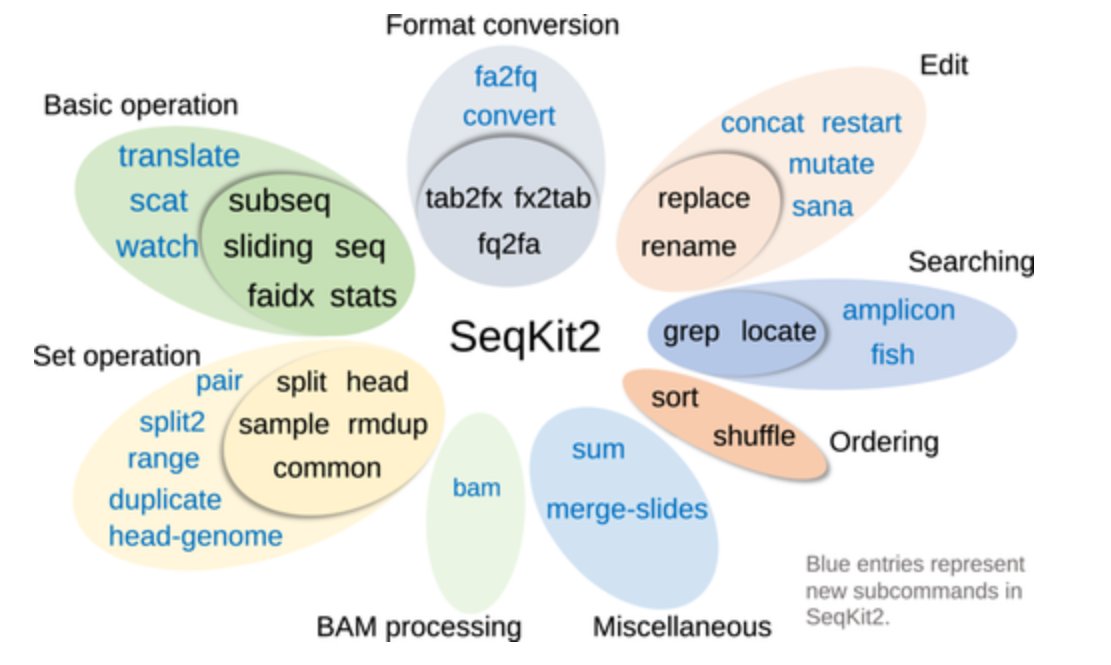
SeqKit is an absolutely amazing tool! I was writing a custom script to chunk reorder the records in a large reference file, and then chunk them into sub-files of a pre-specified size. The script wasn't huge, but was slow & error prone. Alternatively; it's just 2 seqkit commands!
SeqKit2: A Swiss army knife for sequence and alignment processing. It suit for nanopore data! #nanopore doi.org/10.1002/imt2.1…
Minimap2-2.27 released with new presets for accurate Nanopore reads and Illumina Complete Long Reads (both recommended by vendors), along with a few minor features and bug fixes. Output alignment identical to v2.26 except the value at a custom tag. github.com/lh3/minimap2/r…
I have 3 invite codes to the site where the sky is blue. Let me know if you want one.
Very happy to see simpleaf published! @DongzeHe and I are working on some exciting improvements and expansions that I'm looking forward to sharing more about soon :).
simpleaf : A simple, flexible, and scalable framework for single-cell data processing using alevin-fry. #SingleCell #DataProcessing #Rust #Bioinformatics @nomad421 academic.oup.com/bioinformatics…
I got featured in the Fall 2023 edition of the Harvard Public Health Magazine!

Do you use pysam? Currently #pysam builds wheels back to Python 3.6, which has been end-of-life for a couple of years. Please let us know via this survey which Python versions you use and would like pysam to continue supporting. github.com/pysam-develope…
Gotta say I'm feeling this at the moment. The downside to making conda packages out of anything that could prove useful is that I end up looking at the code and... yeah... well, what Nils said.
Dear academics, I'm going to eventually read your code. Please feel the appropriate amount of shame from the start about your hard-coded paths, missing required files, and major assumptions about file naming. Your frenemy, industry.
#Genedata collaborates with @AstellasPharma to help confirm the #biosafety of cell therapies using #NGS-based assays within a validated environment. Read more at buff.ly/45XzA7Z #DigitalizingBiopharma #GenedataSelector #Bioinformatics #gmp #Celltherapy

And finally, it is great to have more than one freely available and well suitable tool to get the job done. What matters is that science is reproducible and transparent, and that these properties are strived for as many people as possible. (3/x)
I've said it before and I'll say it again, it's important that you use A workflow manager, not really which one you use. It's vital to get familiar enough with one to be productive and make reproducible and redistributable analyses. The exact choice is then totally "you do you".
I see tweetdeck is now disabled for us normies. Nothing like making this site even less functional.
TIL that a pie chart is referred to as a Camembert in French and I think we should absolutely call it this in English now!
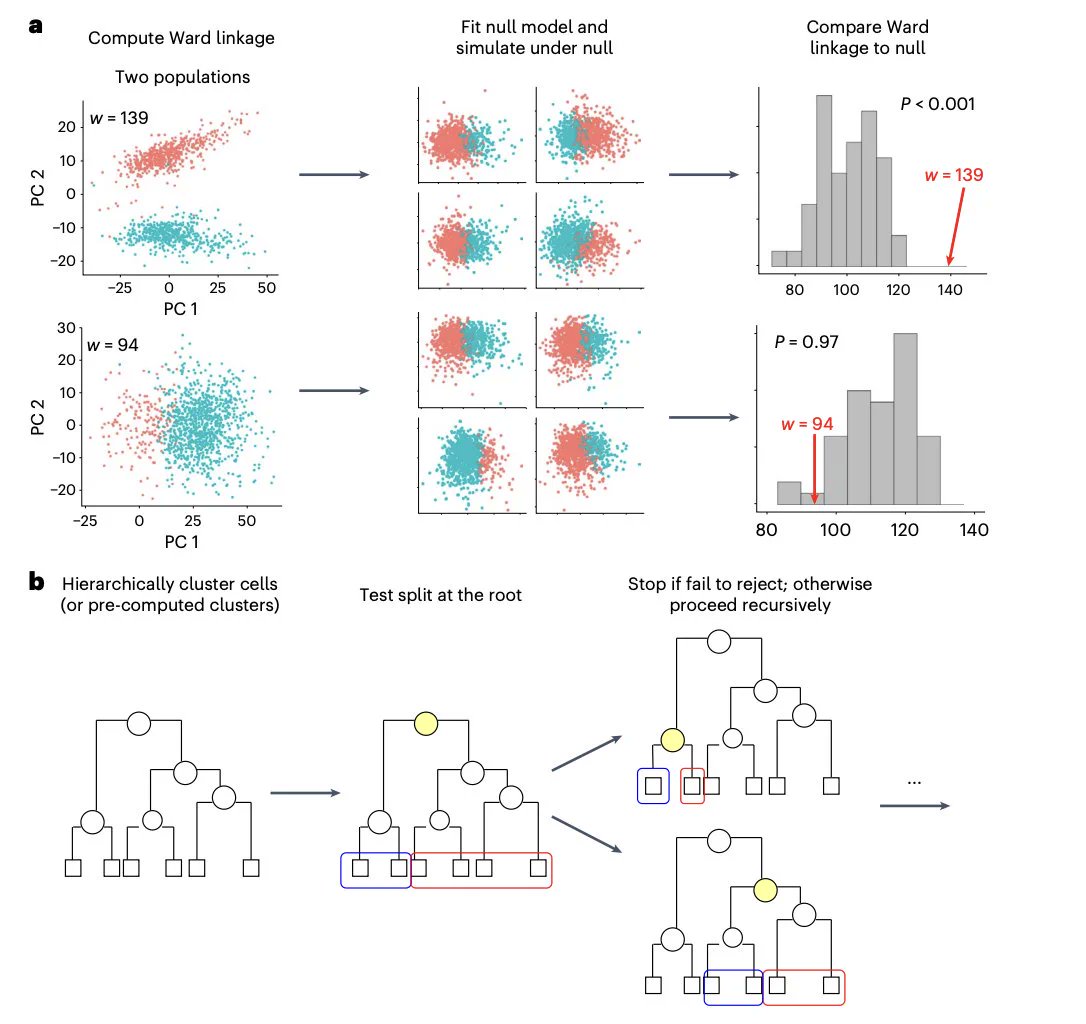
Clustering algorithms report clusters even when none exist. In single-cell RNA-Seq pipelines, novel cell types are often identified by clustering algorithms. Expanding on Kimes et al.'s work, we introduce significance analysis for single-cell RNA-Seq data: nature.com/articles/s4159…
bedder-tools 😜 is the next generation of bedtools from @aaronquinlan lab. It's early days, but we think the foundation is solid. We're looking for feedback on the rust code. Have a look: github.com/quinlan-lab/be… and specifically about design here: github.com/quinlan-lab/be…
Pangene is a new tool for constructing a pangenome gene graph from multiple assemblies: github.com/lh3/pangene. You can explore the rich haplotypic diversity of human genes at pangene.liheng.org. Still WIP.
United States Trends
- 1. Spotify 1,91 Mn posts
- 2. $TOAD 4.437 posts
- 3. $PHNIX 5.903 posts
- 4. Pete 793 B posts
- 5. Brian Thompson 99,1 B posts
- 6. Isak 17,9 B posts
- 7. CEOs 16,7 B posts
- 8. Apple Music 227 B posts
- 9. United Healthcare 73,6 B posts
- 10. Diontae Johnson 2.862 posts
- 11. NASA 77,7 B posts
- 12. Subsonic 3.468 posts
- 13. Newcastle 38,5 B posts
- 14. Chipotle 7.344 posts
- 15. Supreme Court 131 B posts
- 16. Powell 23,2 B posts
- 17. #NEWLIV 14,8 B posts
- 18. ACLU 43,3 B posts
- 19. #DragRace 7.349 posts
- 20. Southampton 31,3 B posts
Who to follow
-
 Johannes Köster (@johanneskoester.bsky.social)
Johannes Köster (@johanneskoester.bsky.social)
@johanneskoester -
 nf-core
nf-core
@nf_core -
 bioRxiv Bioinfo
bioRxiv Bioinfo
@biorxiv_bioinfo -
 Torsten Seemann
Torsten Seemann
@torstenseemann -
Steven Salzberg 💙💛
@StevenSalzberg1 -
 Oliver Stegle
Oliver Stegle
@OliverStegle -
 Erik Garrison
Erik Garrison
@erikgarrison -
 𝕐 (@[email protected], @robp.bsky.social)
𝕐 (@[email protected], @robp.bsky.social)
@nomad421 -
 Alicia Oshlack
Alicia Oshlack
@AliciaOshlack -
 Mark R@markrobinsonca@{genomic.social,bsky.social}
Mark R@markrobinsonca@{genomic.social,bsky.social}
@markrobinsonca -
 Michael Schatz
Michael Schatz
@mike_schatz -
 Wolfgang Huber 🇺🇦
Wolfgang Huber 🇺🇦
@wolfgangkhuber -
 brent pedersen
brent pedersen
@brent_p -
 Paolo Di Tommaso
Paolo Di Tommaso
@PaoloDiTommaso -
 Rayan Chikhi
Rayan Chikhi
@RayanChikhi
Something went wrong.
Something went wrong.