Dina Hochhauser
@dinahochPhD student @SorekLab @WeizmannScience studying phage-bacteria interactions 🦠, MGEs 🛒 & defence islands 🛡🏝 @dinahoch.bsky.social
Similar User

@AudeBer

@SorekLab

@MDMlab_Paris

@rafapinilla92

@mfwhite2

@SarahCamara94

@AAvihail

@IngaTrails

@PeterFineran

@AShaidullina1

@BursteinLab

@epcrocha

@NobregaFL

@FrancoisRousset

@FrunzkeLab
It’s OUT! 🥳 Beyond excited to see my work in the @SorekLab in print! 🤩 The defense island repertoire of the Escherichia coli pan-genome journals.plos.org/plosgenetics/a… Yes, I had to change the spelling from ‘defence’ 🇬🇧 to ‘defense’ 🇺🇸, but we have some new results too! ⬇️
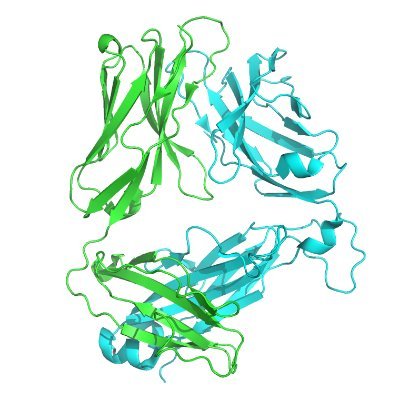
AcrIIIA1 is a protein–RNA anti-CRISPR complex that targets core Cas and accessory nucleases academic.oup.com/nar/article/do…
It has been a journey and here it is: my first post doc paper is out in Nature Communications 🎉🎉🎉 If you red the preprint, it's worth checking the revised manuscript, we added some cool stuff. A thread. ⬇️ nature.com/articles/s4146…
Published today @NatureComms We discovered a bacterial NLR-like pattern recognition receptor that can sense 3 different viral proteins as a signature for infection. This explains its broad defense against different phage families Congrats @nath_bechon! nature.com/articles/s4146…
New GPU-based MMseqs2: 20x faster searches on a single L40S (approx. as fast as a RTX 4090) vs. a 128-core CPU. This work enables to set up a very cost-efficient ColabFold MSA GPU server. 🧵 📄biorxiv.org/content/10.110… 💾mmseqs.com 🗞️ developer.nvidia.com/blog/boost-alp…

Telomeric transposons are pervasive in linear bacterial genomes biorxiv.org/cgi/content/sh… #biorxiv_micrbio
Accelerated computing is powering a new era in protein structure prediction, now with speed-of-light multiple sequence alignments. #MMseqs2GPU now makes #AlphaFold2 predictions faster and more efficient than ever. 1/🧵 developer.nvidia.com/blog/boost-alp…
I’m excited to share the first preprint from my postdoc at @johan_elf_ lab together with Konrad Gras. 🧬🦠 biorxiv.org/content/10.110… (1/14)
Pre-print 🚨 alert! We used AI protein structure prediction to analyze ~10,000 Borg proteins, revealing potential capsid proteins & viral-like features in these massive elements from methane-eating archaea. Could they be the first archaeal giant viruses? Check it out!

For the past two years, we have been working on a novel approach to identify origins of transfer by conjugation in plasmids from any bacterial species. We are thrilled to see this work now published in @NatureMicrobiol after thorough peer review! nature.com/articles/s4156…
Excited to share that the AlphaFold 3 model code and weights are now available for academic use. Looking forward to seeing what new research this unlocks and how the research community builds on AlphaFold 3 for scientific discoveries github.com/google-deepmin… 1/2
Expanding the diversity of origin of transfer-containing sequences in mobilizable plasmids | Nature Microbiology idp.nature.com/authorize?resp…
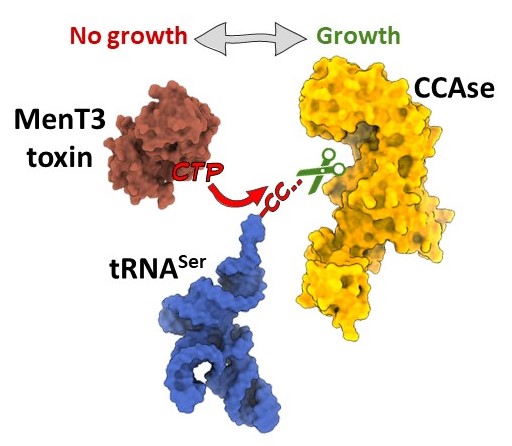
🙏Our latest paper on MenT3 in vivo with @xibing4, @BlowerLab, @ONeyrolles rdcu.be/dZle4 MenT3 targets serine tRNA in vivo/MenT3 is active under standard growth condition/The CCAse PcnA counteracts MenT3 activity. @FRM_officiel , @AgenceRecherche, @CbiToulouse
Activation of a helper NLR by plant and bacterial TIR immune signaling science.org/doi/10.1126/sc…
Bacterial warfare is associated with virulence and antimicrobial resistance biorxiv.org/cgi/content/sh… #biorxiv_micrbio
biorxiv.org/content/10.110… BORGS - the large extrachromosomal elements of anaerobic methane-oxidizing archaea are likely viruses and have evolved genomic features similar to Giant Viruses!
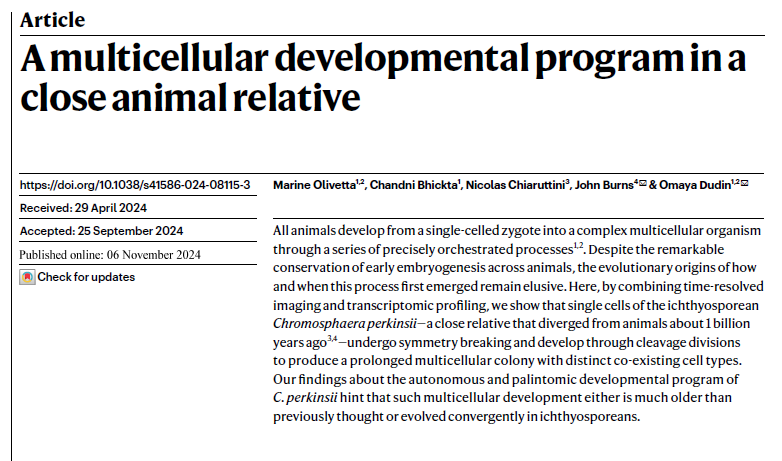
@dudin_o's lab just released a groundbreaking paper showing that one of animals' closest unicellular relatives has a developmental program resembling animal embryos (image from @thibaut_brunet)! This fundamentally changes how I think about the origin of animal development. 1/8

Our latest #Ichthyosporean #EvoDevo story, led by M. Olivetta @ChandniBhickta @NChiaruttini & in co. with @burnsajohn is finally out @Nature . Link⏩: nature.com/articles/s4158… Preprint⏩: biorxiv.org/content/10.110… 1/5)
OUT NOW A widespread phage-encoded kinase enables evasion of multiple host antiphage defense systems nature.com/articles/s4156…
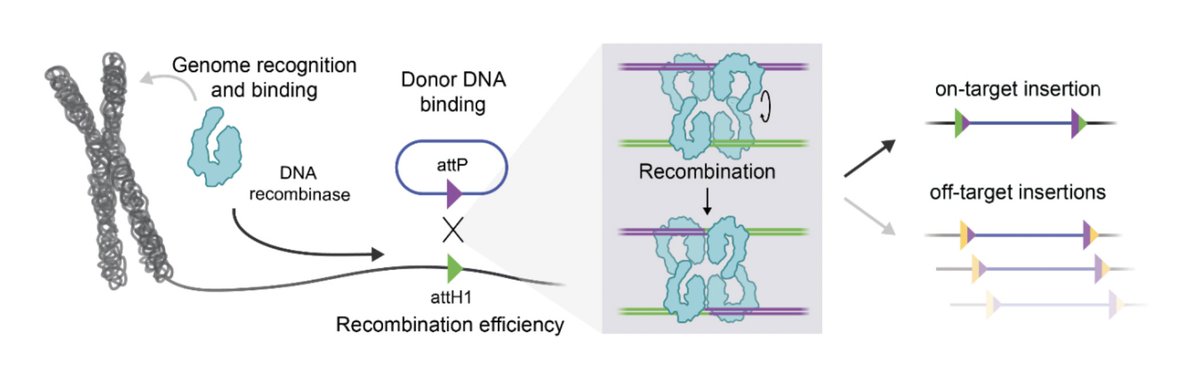
🧵In new work, we report a systematic engineering roadmap to optimize large serine recombinases (LSRs) for direct, site-specific insertion into the human genome 🧬. We achieved over 50% insertion efficiency and 97% genome-wide specificity, a 10X improvement over our previous work
🧬🧬 𝙉𝙚𝙬 𝙋𝙧𝙚𝙥𝙧𝙞𝙣𝙩!!🧬🧬 Latest collab with the amazing Ming Li & Yue Feng labs! How do phages bypass multi-layered defenses in bacteria? Spoiler: with equally complex anti-defenses!! 🧵👇 biorxiv.org/content/10.110…
Nature News piece on the hidden players of the gut microbiome (hint, they're phages, of course) “Our virome is hugely abundant and incredibly diverse, and we’ve looked at just a tiny percentage,” nature.com/articles/d4158…
United States Trends
- 1. Celtics 22 B posts
- 2. Cavs 21,2 B posts
- 3. #OnlyKash 71,4 B posts
- 4. #MCADE N/A
- 5. Pat Murphy 2.258 posts
- 6. Joey Galloway N/A
- 7. Nancy Mace 96,2 B posts
- 8. Mendoza 9.688 posts
- 9. Cenk 18,3 B posts
- 10. Jaguar 69,3 B posts
- 11. Mobley 2.300 posts
- 12. Linda McMahon 6.780 posts
- 13. Queta N/A
- 14. Medicare and Medicaid 30,9 B posts
- 15. Starship 215 B posts
- 16. #spitemoney N/A
- 17. College Football Playoff 3.674 posts
- 18. SpaceX 232 B posts
- 19. Lichtman 3.393 posts
- 20. Sweeney 12,5 B posts
Who to follow
-
 Aude Bernheim
Aude Bernheim
@AudeBer -
 Sorek Lab
Sorek Lab
@SorekLab -
 MDMLab
MDMLab
@MDMlab_Paris -
 Rafa Pinilla-Redondo
Rafa Pinilla-Redondo
@rafapinilla92 -
 Malcolm White
Malcolm White
@mfwhite2 -
 Sarah Camara
Sarah Camara
@SarahCamara94 -
 Avigail Stokar-Avihail
Avigail Stokar-Avihail
@AAvihail -
 Inga Songailiene
Inga Songailiene
@IngaTrails -
 Peter Fineran
Peter Fineran
@PeterFineran -
 Aisylu Shaidullina
Aisylu Shaidullina
@AShaidullina1 -
 Burstein lab
Burstein lab
@BursteinLab -
 Eduardo Rocha
Eduardo Rocha
@epcrocha -
 Franklin Nobrega
Franklin Nobrega
@NobregaFL -
 François Rousset
François Rousset
@FrancoisRousset -
 FrunzkeLab
FrunzkeLab
@FrunzkeLab
Something went wrong.
Something went wrong.