Similar User

@NorbertPiske

@alexanderfuxi

@bhagirathl

@RuochiZhang

@psimslab

@DrRonaldMa1

@GersteinLab

@DouglasYaoDY

@Hongbo919Liu

@_SteveGiguere_

@Z_Linying

@HY3952

@karbalayghareh

@SunSherine

@dticoll
Commonly used software tools produce conflicting and overly-optimistic AUPRC values genomebiology.biomedcentral.com/articles/10.11… 🧬🖥️ github.com/wychencuhk/AUP…
Now on Genome Biology: genomebiology.biomedcentral.com/articles/10.11…
1/6 Are you really winning? 🤔️ We dive into 10 ML toolkits used in 3K+ papers and discovered some🤯 facts: ❌ Different tools give different AUPRC 🚨 Only 2/10 tools correctly implemented AUPRC 👀 Winners of ML contests ($$$) changed if using different tools See more in 🧵:
1/6 Are you really winning? 🤔️ We dive into 10 ML toolkits used in 3K+ papers and discovered some🤯 facts: ❌ Different tools give different AUPRC 🚨 Only 2/10 tools correctly implemented AUPRC 👀 Winners of ML contests ($$$) changed if using different tools See more in 🧵:
Commonly used software tools produce conflicting and overly-optimistic AUPRC values biorxiv.org/cgi/content/sh… #bioRxiv
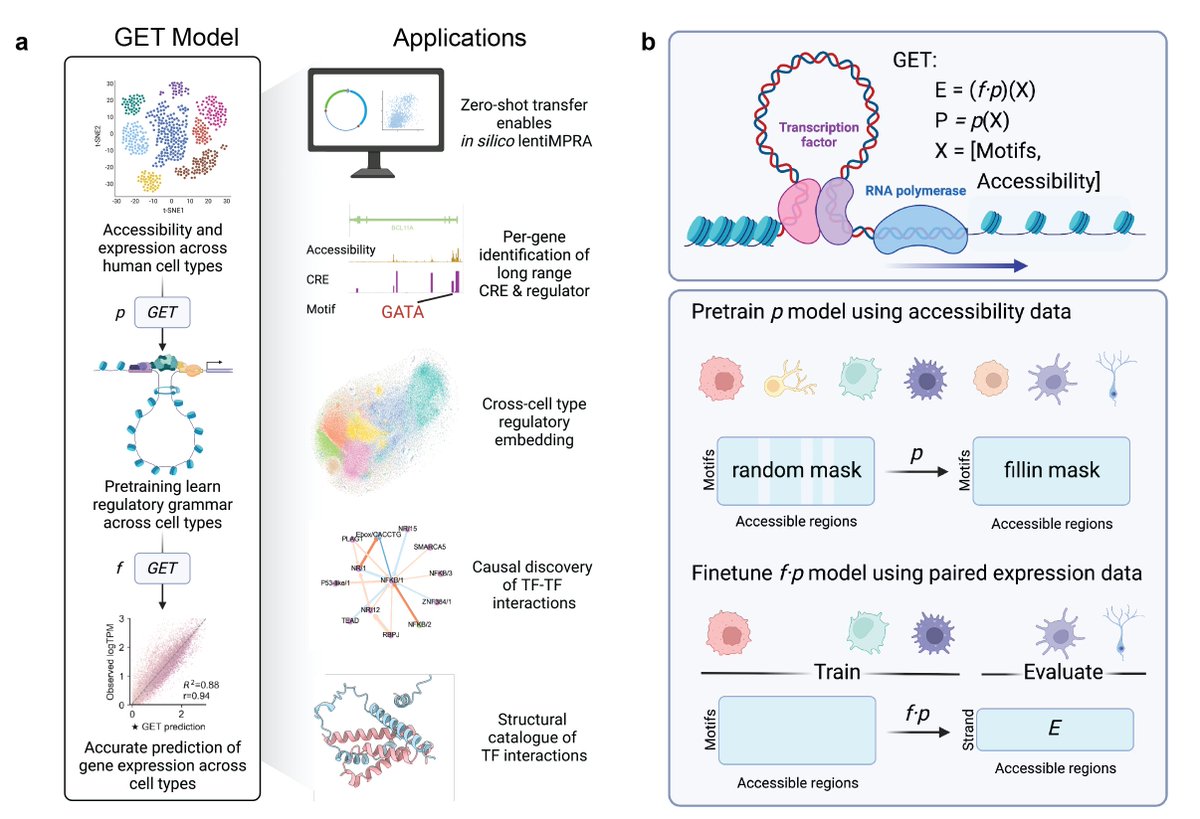
Glad to share our preprint "GET: a foundation model of transcription across human cell types" biorxiv.org/content/10.110… A interactive demo (beta) for regulatory analysis and structure catalog of transcription factor interactions can be viewed at huggingface.co/spaces/get-fou…
Commonly used software tools produce conflicting and overly-optimistic AUPRC values biorxiv.org/cgi/content/sh… #biorxiv_bioinfo
When I saw topologically associating domains (TADs) drawn as globular structures in review papers, I always wondered whether TADs really have functionally important 3D structures. If you also wonder about it, check out our new preprint: doi.org/10.1101/2022.0…
Graph neural networks can capture complex object relationships. Eg, they can identify cancer genes based on omic features and protein interactions. See our new NMI paper rdcu.be/cJcs4 by @xxmen21 and @cqhoneybear to find out the tricks!
👏👏👏
I'm glad to share a thread on my paper studying disease-associating noncoding regulatory regions, just published on @genomeresearch November issue: genome.cshlp.org/content/30/11/… (together with an unpublished sci-fi theme cover art 🤣)

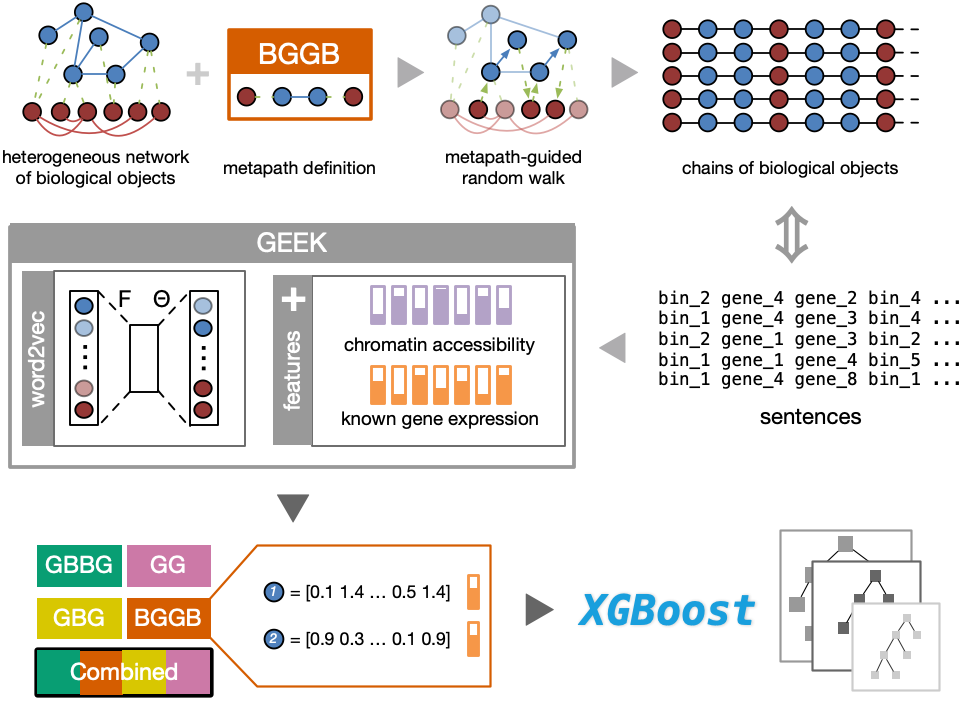
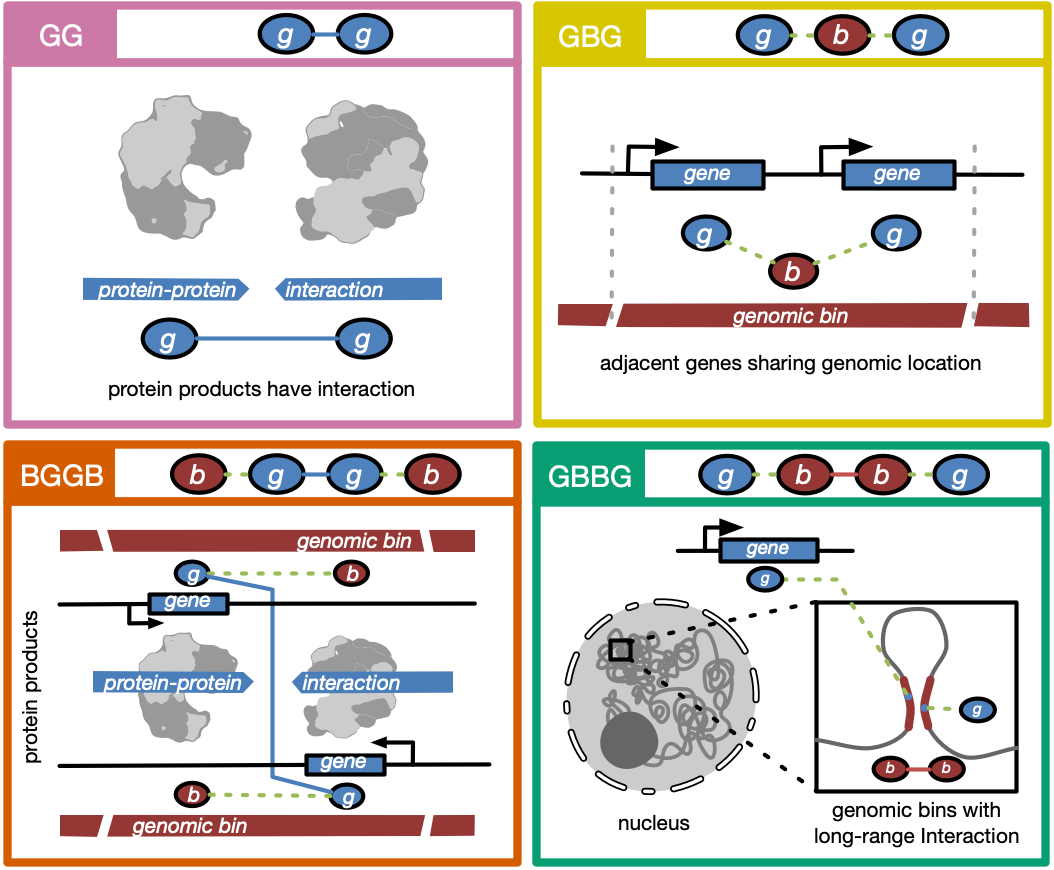
Happy to share you our freshly published work on @NatMachIntell We proposed GEEK, a flexible, network-embedding-based framework integrating heterogeneous biological knowledge to study their joint effects on gene expression. Read full text: rdcu.be/b5R48
Super excited to share our new framework for the analysis of #noncoding #variants in #WGS! -> Whole-genome analysis of noncoding genetic variations identifies multigranular regulatory element perturbations with Hirschsprung disease disq.us/t/3nn5k18
HiDRA: Building on Sharpr-MPRA+STARR-Seq+ATAC-Seq to dissect millions of human regulatory regions in one experiment biorxiv.org/content/early/…




Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter… dlvr.it/Ppvl49
United States Trends
- 1. #Survivor47 3.579 posts
- 2. #AEWDynamite 11 B posts
- 3. Tyler Herro 1.508 posts
- 4. Fauci 125 B posts
- 5. #AbbottElementary 2.733 posts
- 6. Spotify 2,92 Mn posts
- 7. Snape 6.683 posts
- 8. #TheMaskedSinger 1.714 posts
- 9. Genevieve 2.103 posts
- 10. #SistasOnBET N/A
- 11. Hawk Tuah 28,4 B posts
- 12. CEOs 28,2 B posts
- 13. Lady Vols N/A
- 14. Teeny 2.909 posts
- 15. $HAWK 10 B posts
- 16. Pete 952 B posts
- 17. Hubert 4.052 posts
- 18. Anthony Davis 1.741 posts
- 19. Ian Jackson N/A
- 20. Cornyn 21,6 B posts
Who to follow
-
 Norbert Piske
Norbert Piske
@NorbertPiske -
 Xi Fu
Xi Fu
@alexanderfuxi -
 Bʜᴀɢɪʀᴀᴛʜ Kᴜᴍᴀʀ Lᴀᴅᴇʀ 🌴🐅🐘🦒🦌
Bʜᴀɢɪʀᴀᴛʜ Kᴜᴍᴀʀ Lᴀᴅᴇʀ 🌴🐅🐘🦒🦌
@bhagirathl -
 Ruochi Zhang
Ruochi Zhang
@RuochiZhang -
 Peter Sims
Peter Sims
@psimslab -
 Dr Ronald Ma
Dr Ronald Ma
@DrRonaldMa1 -
 Gerstein Lab | Yale
Gerstein Lab | Yale
@GersteinLab -
 Douglas Yao
Douglas Yao
@DouglasYaoDY -
 Hongbo Liu
Hongbo Liu
@Hongbo919Liu -
 Steve Giguere
Steve Giguere
@_SteveGiguere_ -
 Linying Zhang
Linying Zhang
@Z_Linying -
 Han Yuan
Han Yuan
@HY3952 -
 Alireza Karbalayghareh
Alireza Karbalayghareh
@karbalayghareh -
 Xueqin (Sherine) Sun
Xueqin (Sherine) Sun
@SunSherine -
 David Ticoll
David Ticoll
@dticoll
Something went wrong.
Something went wrong.