Zhenghao Zhang
@adam_zhzhangPhD @ CUHK CSE, Bioinformatics, Single-cell DS, Multi-omics
Similar User

@chensj16

@alexanderfuxi

@debdatta_halder

@johnding86

@DormantSideKick

@AAHaider0006

@OierAzur
Commonly used software tools produce conflicting and overly-optimistic AUPRC values genomebiology.biomedcentral.com/articles/10.11… 🧬🖥️ github.com/wychencuhk/AUP…
Now on Genome Biology: genomebiology.biomedcentral.com/articles/10.11…
1/6 Are you really winning? 🤔️ We dive into 10 ML toolkits used in 3K+ papers and discovered some🤯 facts: ❌ Different tools give different AUPRC 🚨 Only 2/10 tools correctly implemented AUPRC 👀 Winners of ML contests ($$$) changed if using different tools See more in 🧵:
Happy to share that I've survived the PhD defense! I couldn't have done it without the incredible support of my supervisor @KevinYipLab, my amazing collaborators (esp. @cqhoneybear), my loving family, and frens who kept me sane during this wild ride. Thanks for being my rock! 🎓
Commonly used software tools produce conflicting and overly-optimistic AUPRC values biorxiv.org/cgi/content/sh… #biorxiv_bioinfo
Commonly used software tools produce conflicting and overly-optimistic AUPRC values biorxiv.org/cgi/content/sh… #bioRxiv
Large Scale Foundation Model on Single-cell Transcriptomics biorxiv.org/cgi/content/sh… #bioRxiv
Epic cover art
I'm glad to share a thread on my paper studying disease-associating noncoding regulatory regions, just published on @genomeresearch November issue: genome.cshlp.org/content/30/11/… (together with an unpublished sci-fi theme cover art 🤣)

MARVEL uses @nextflowio in its core and embraces the @Docker trend, so it should be relatively easy for you to apply it to your own WGS study or scale it up. Try it here: github.com/fuxialexander/…
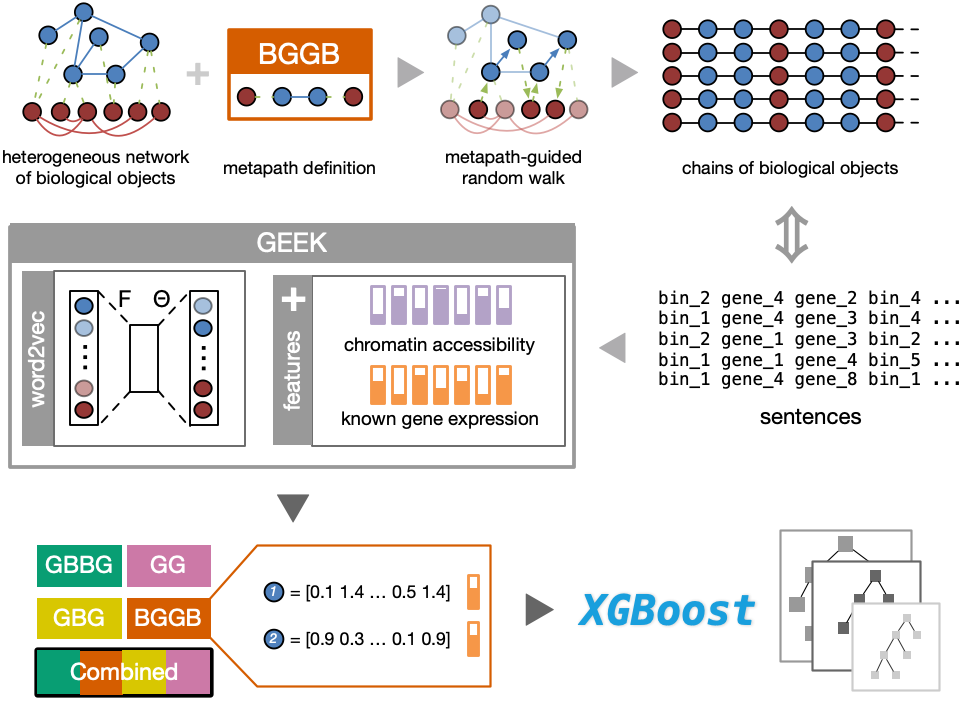
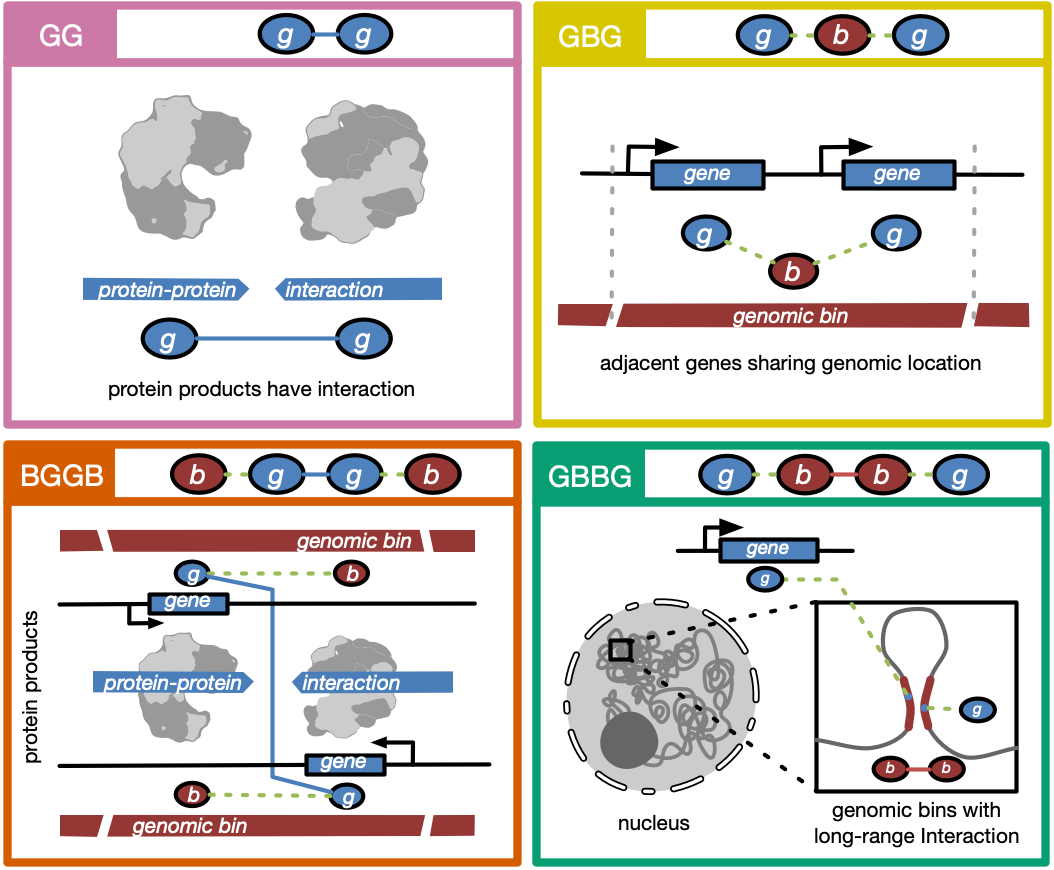
How to integrate #multiomics biological data (chromatin interaction, accessibility, expression, etc ) in a heterogeneous network? #metapath and network #embedding could be helpful. Checkout GEEK, a new framework developed by my groupmates Qin and Adam @adam_zhzhang at CUHK!
Happy to share you our freshly published work on @NatMachIntell We proposed GEEK, a flexible, network-embedding-based framework integrating heterogeneous biological knowledge to study their joint effects on gene expression. Read full text: rdcu.be/b5R48
Super excited to share our new framework for the analysis of #noncoding #variants in #WGS! -> Whole-genome analysis of noncoding genetic variations identifies multigranular regulatory element perturbations with Hirschsprung disease disq.us/t/3nn5k18
United States Trends
- 1. Lakers 27,3 B posts
- 2. #Survivor47 5.629 posts
- 3. #AEWDynamite 14,6 B posts
- 4. Tyler Herro 2.804 posts
- 5. Fauci 133 B posts
- 6. #AbbottElementary 2.948 posts
- 7. #SistasOnBET 1.190 posts
- 8. Anthony Davis 2.262 posts
- 9. Genevieve 2.584 posts
- 10. Operation Italy 1.846 posts
- 11. Miles Kelly N/A
- 12. #TheChallenge40 N/A
- 13. Hawk Tuah 29,7 B posts
- 14. Hubert 4.360 posts
- 15. Teeny 3.239 posts
- 16. Snape 7.202 posts
- 17. Creighton 2.884 posts
- 18. CEOs 29,2 B posts
- 19. Lady Vols N/A
- 20. $HAWK 10,8 B posts
Something went wrong.
Something went wrong.