Yuheng Fu
@cadyyuhengPhD Student @Yi_Lab_NU @Rosemary_Braun | CompBio Enthusiast @NUFeinbergMed | WashU '20 @morris_lab
Similar User

@kenjikamimoto68

@junedh_amrute

@Jeongmini_L

@dgpnorthwestern

@kjkjindal

@Ruli_Gao

@arpandas71

@ChaudhuriShuvam

@HaikuoLi

@EBartom

@leung_holden
scHolography is online! It enables single-cell spatial neighborhood reconstruction, analysis, and visualization. Big thanks to the team, @arpandas71 and Dongmei, and to the guidance of my advisors @Yi_Lab_NU and @Rosemary_Braun @LurieCancer @NU_Pathology doi.org/10.1186/s13059…
We have open positions for postdocs or staff scientists. My lab will study the developmental process and biomedical engineering by integrating single-cell omics, informatics, and predictive modeling. Please contact me if you are looking for a position!! Please RT!
Some exciting news to share! Our lab is relocating to @harvardmed Systems Biology and @BrighamWomens GI & Genetics this fall. We're actively recruiting talented postdocs to join our team. Please RT to spread the word.

Review: We summarize recent advances in machine learning for multi-omics integration, and limitations in current methodologies. We propose a new biology-inspired AI framework for multi-omics integration and predictive modeling of human phenotypic responses to novel perturbations
AI-driven multi-omics integration for multi-scale predictive modeling of causal genotype-environment-phenotype relationships. arxiv.org/abs/2407.06405
An article published in @GenomeBiology presents scHolography: a machine learning-based method designed to reconstruct single-cell spatial neighborhoods and facilitate 3D tissue visualization using spatial and single-cell RNA sequencing data. genomebiology.biomedcentral.com/articles/10.11…
scHolography: a computational method for single-cell spatial neighborhood reconstruction and analysis genomebiology.biomedcentral.com/articles/10.11…

Very excited to share our newest tool: scHolography for reconstruction and quantitative analysis of spatial neighborhoods. Fantastic work by @cadyyuheng and collaboration with @Rosemary_Braun Visit github.com/YiLab-SC/scHol… for tutorials. genomebiology.biomedcentral.com/articles/10.11…
Our paper characterizing the impact of different #geneexpression normalization with different gene panels in analysis + interpretation of #singlecell imaging-based targeted #spatialtranscriptomics data is now out @GenomeBiology Summary and 🧵👇 Congrats to @lylaatta + team 🥂

Check out our now published investigation into how experimental design and analysis choices can affect downstream results when analyzing imaging-based spatial transcriptomics data. rdcu.be/dKFby
Inspired by @lpachter and team's exploration of scRNA-seq pipelines, we face similar challenges in scATAC-seq, only magnified! After 5 years of tackling these inconsistencies with my advisor @JunhyongKim , Here is a summary (and some solutions😃) 1/n #Genomics #Bioinformatics
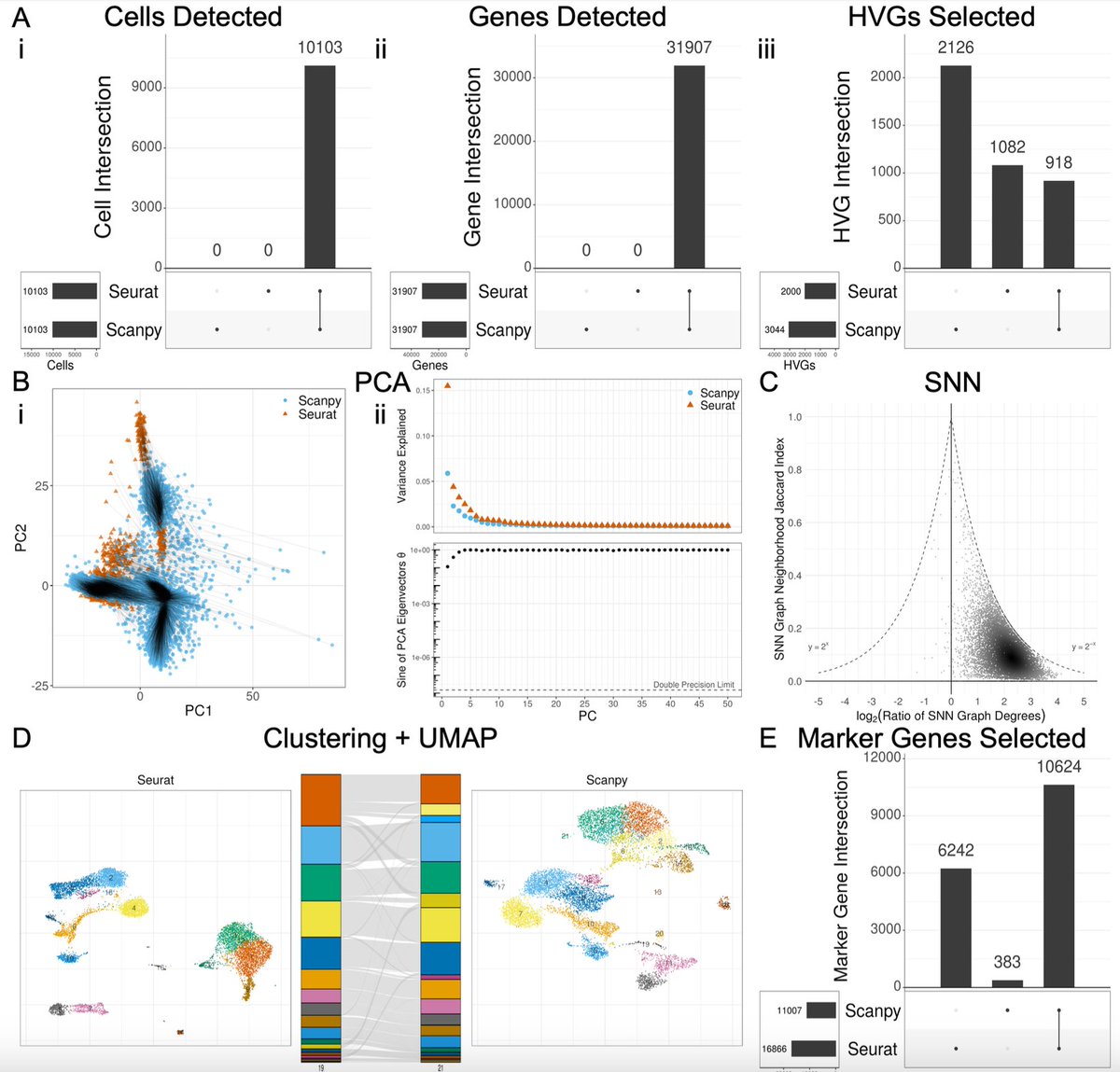
The choice of whether to use Seurat or Scanpy for single-cell RNA-seq analysis typically comes down to a preference of R vs. Python. But do they produce the same results? In biorxiv.org/content/10.110… w/ @Josephmrich et al. we take a close look. The results are 👀 1/🧵
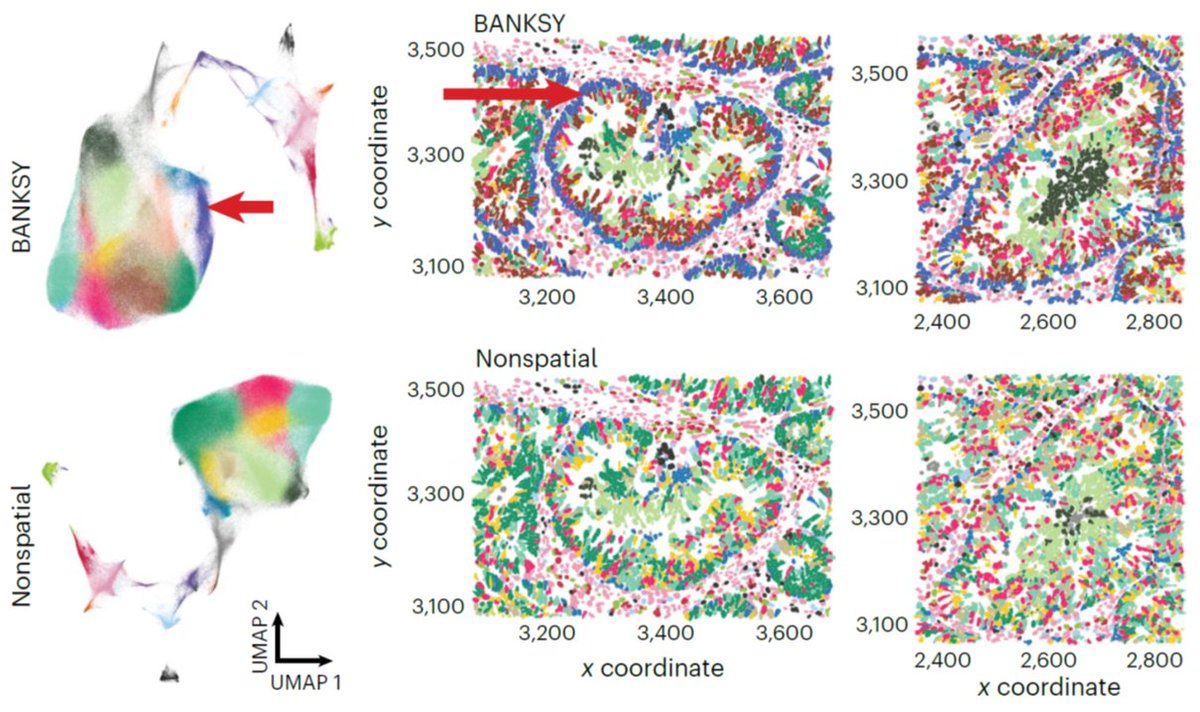
1. BANKSY is out! go.nature.com/4bUR200 Our spatial clustering algo applies to any spatial RNA/protein/... assay, scales to 2M cells, detects both cell typing and spatial domains and facilitates spatial batch correction. 🧵 👇
We introduce Epiregulon, a method to infer transcription factor activity at the single cell level, with the goal of identifying master regulators of disease and drug response. Preprint: biorxiv.org/content/10.110… GitHub: github.com/xiaosaiyao/epi…
A detailed review on bipartite graphs in systems biology and medicine: a survey of methods and applications doi.org/10.1093/gigasc… #sysbio

UMAP and t-SNE are widely used in single-cell genomics to identifying features of interest, and visually explore data. In a new paper w/ Tara Chari we find that extensive distortions and inconsistent practices make such embeddings counter-productive.🧵journals.plos.org/ploscompbiol/a… 1/
Our single-cell RNA-seq and ATAC-seq read simulator scReadSim is now online at @NatureComms nature.com/articles/s4146… scReadSim mimics real data at read-sequence and read-count levels, and it provides ground truths: UMI counts for RNA-seq and open chromatin regions for ATAC. 1/
WGCNA is cited 16,372 times for bulk RNAseq co-expression analysis. The single-cell version is here: hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data buff.ly/47dMTl2 My blog post on it a while ago buff.ly/49nyTqA
Our multi-modal single-cell skin atlas is now online @biorxivpreprint Co-led by @gokcen, Maria Alora-Palli, Raif Geha, #AvivRegev, @TheXavierLab biorxiv.org/content/10.110…
A human prenatal skin cell atlas reveals immune cell regulation of skin morphogenesis biorxiv.org/cgi/content/sh… #bioRxiv
Online now: Molecular and spatial landmarks of early mouse skin development dlvr.it/StkgXr
(1) This is an amazing paper on the limitations of (current) single-cell foundation models "Our results indicate that both Geneformer and scGPT exhibit limited reliability in zero-shot settings and often underperform compared to simpler methods." biorxiv.org/content/10.110…

StrastiveVI: Isolating structured salient variations in single-cell transcriptomic data biorxiv.org/content/10.110…

United States Trends
- 1. Travis Hunter 16,4 B posts
- 2. Clemson 7.556 posts
- 3. Colorado 71,6 B posts
- 4. Arkansas 28,4 B posts
- 5. Dabo 1.367 posts
- 6. Quinn 14,9 B posts
- 7. #SkoBuffs 4.462 posts
- 8. Cam Coleman 1.092 posts
- 9. Isaac Wilson N/A
- 10. #HookEm 3.263 posts
- 11. Zepeda 2.018 posts
- 12. Sean McDonough N/A
- 13. $CUTO 8.287 posts
- 14. Northwestern 6.867 posts
- 15. Tulane 2.880 posts
- 16. #NWSL N/A
- 17. Sark 1.978 posts
- 18. #iubb N/A
- 19. Pentagon 98,4 B posts
- 20. Mercer 4.322 posts
Who to follow
-
 Kenji Kamimoto
Kenji Kamimoto
@kenjikamimoto68 -
 Junedh M. Amrute
Junedh M. Amrute
@junedh_amrute -
 Jeongmin Lee 🧬🧫
Jeongmin Lee 🧬🧫
@Jeongmini_L -
 Driskill Graduate Program in Life Sci
Driskill Graduate Program in Life Sci
@dgpnorthwestern -
 Kunal Jindal
Kunal Jindal
@kjkjindal -
 Ruli_Gao
Ruli_Gao
@Ruli_Gao -
 Arpan Das 😷
Arpan Das 😷
@arpandas71 -
 Shuvam Chaudhuri
Shuvam Chaudhuri
@ChaudhuriShuvam -
 Haikuo Li
Haikuo Li
@HaikuoLi -
 Elizabeth Bartom
Elizabeth Bartom
@EBartom -
 Yonghao Liang
Yonghao Liang
@leung_holden
Something went wrong.
Something went wrong.