Armin Scheben
@arminschebenPostdoc in Computational Biology at Cold Spring Harbor Laboratory. Doing comparative genomics, mostly in🌱. Sharing cool #science.
Similar User

@UllasPedmale

@chirgjain

@jmkreiner

@MaizeZynskiHI

@JinliangYang

@SigmaFacto

@toheitka

@CarcySalome

@lakeseafly1

@KeithLAdams

@jrossibarra

@skimomiks

@aylwyn_scally

@yongfeng_zhou

@MGStetter
New genetic barcoding technology developed at @WeillCornell can help CSHL researchers map where metastatic prostate cancer spreads, and maybe one day roadblock cancer. @asiepel @arminscheben cshl.edu/how-does-cance…
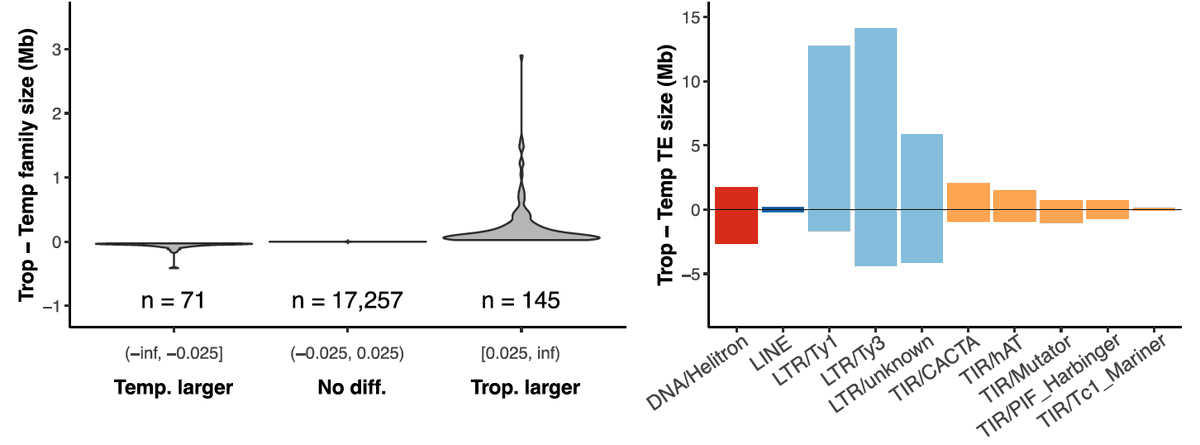
How TEs gain and lose between tropical and temperate maize inbreds? We found that tropical corns have 35Mb more TEs than temperates, and it's mainly due to a small number of LTR families that are young, less methylated, and more expressed in tropical maize.pubmed.ncbi.nlm.nih.gov/39251347/
PlantCaduceus - Cross-species modeling of plant genomes Led by Jingjing Zhai and Aaron Gokaslan, with the Kuleshov lab, we’re having our “ChatGPT moment” with DNA Large Models. PlantCaduceus predicts functional DNA sequences with single base resolution across flowering plants.
So great to see this paper out in @Nature Beautiful molecular characterization of a new toxin-antidote drive system in maize, led by Ben berube, with assistance from Martienssen lab @antnerves @CrisAlves_bio @BioJlynn and collaborators @arminscheben @jrossibarra Jerry Kermicle
Why was corn able to spread and adapt so rapidly across the Americas 4,000 years ago? Research from CSHL’s Rob Martienssen suggests that the answer may lie in a genetic system called Teosinte Pollen Drive. cshl.edu/corns-missing-…
Very excited to be starting my group this autumn at ithems.riken.jp/en @RIKEN_JP and we’re hiring! riken.jp/en/careers/res… Many more fellowship opportunities as well, please get in touch if interested and see you in Tokyo!
My group is recruiting a bioinformatician for genomic studies in maize. The position includes work on diverse projects and collaborations with the Maize Genetics and Genomics Database (@MaizeGDB), the CERCA project, and others in the Hufford Lab. isu.wd1.myworkdayjobs.com/IowaStateJobs/…
You like plant biodiversity and genomic methods? We are looking for a #Postdoc @LMU_Muenchen Apply until July 31, 2024. Please RT! 🌱🌵🧬#PlantSciJobs job-portal.lmu.de/jobposting/d48…
Had an invigorating day of scientific presentation and discussion at Friday's #NYAPG24 meeting with @Stephen_Stak The poster sessions on a rooftop were a great opportunity to draw in the plant-curious crowd and reconnect with old friends and poster neighbors like @JianjunJin

I am hiring a postdoc to work on evolutionary genomics projects with maize and its wild relatives. This is part of a broad collaborative project known as CERCA that aims to improve maize with adaptations from the wild. Please share or consider applying! isu.wd1.myworkdayjobs.com/IowaStateJobs/…
I'm excited to share our latest research on the mechanisms of splice-modifying drugs. We studied how two small-molecule drugs, risdiplam and branaplam, work. The results challenge the existing understanding of these two drugs. nature.com/articles/s4146…
The MaizeCODE paper is on BioRXiv! New insights on enhancers in plants and a lot of data for every maize (epi)geneticist to play with, including on teosinte, the ancestor of maize. A big team effort, very grateful to everyone involved! biorxiv.org/content/10.110…
MaizeCODE reveals bi-directionally expressed enhancers that harbor molecular signatures of maize domestication. biorxiv.org/cgi/content/sh… #biorxiv_plants
Thread about a new preprint from the lab, led by Logan Whitehouse and Dylan Ray, about tree-sequences. The teaser tagline: why use tree sequences for evolutionary inference when you can directly examine population genetic alignments? biorxiv.org/content/10.110…
Half a million models + many duck jokes later, I'm excited to share the preprint for nQuack: biorxiv.org/content/10.110… This project started as a paragraph in a paper with @JLandisBotany, @RobertLaport, and @desert_hopping But morphed into a new method and package. (1/n)
.@ShellyGaynor introduces nQuack, a program to ID autopolyploids w/sequencing data - many theoretical advantages over similar tools like nQuire. Will be available on GitHub soon! Very grateful for folks like Shelly who do the math so I don’t have to… #Botany2023
Come join us @ZuckermanBrain on May 17th for the 2024 New York Area Population Genetics Meeting. Keynotes by @lindymcbr and @CedricFeschotte Register here zuckermaninstitute.columbia.edu/new-york-area-…
Interested in cutting-edge genomics, pan-genomics, and crop bioinformatics? 🌱💻 Join our newly established Computational Plant Biology group at TUM! We're excited to see your application. Please spread the word! Apply now: shorturl.at/hxBOS #Genomics #Bioinformatics #TUM
The Zeavolution webinar on maize evolution will have it's first 2024 meeting tomorrow at 10am CST. @MohamedElWalid (Buckler Lab at Cornell) will get us started! DM me if you need to be added to the Zeavolution Slack, need connection details, or would like to speak in the series!
Great to return to San Diego for #PAG31 to present my work on constraint and divergence in the noncoding genomes of maize and its relatives in the 'Comparative Genomics' workshop today at 10.30am. Looking forward to lots of conversations on genomics and beyond.


Super excited about our new paper entitled ‘Convergence in carnivorous pitcher plants reveals a mechanism for composite trait evolution’ just out in @ScienceMagazine and featured on the cover!

Happy to share our recent work publishing in a rising journal @iMetaScience . Duck pan‐genome reveals two transposon insertions caused bodyweight enlarging and white plumage phenotype formation during evolution. And thanks for mybro support @arminscheben onlinelibrary.wiley.com/doi/10.1002/im…
United States Trends
- 1. #AskFFT N/A
- 2. Good Sunday 63 B posts
- 3. #sundayvibes 6.714 posts
- 4. Chuck Woolery 10 B posts
- 5. #chillguy 28,3 B posts
- 6. #AskZB N/A
- 7. #ipl2025auction 28 B posts
- 8. Wheel of Fortune 4.597 posts
- 9. Feliz Domingo 13,2 B posts
- 10. Love Connection 5.808 posts
- 11. Ashwin 24,6 B posts
- 12. Lord's Day 2.785 posts
- 13. And the Word 87,9 B posts
- 14. Blessed Sunday 15,9 B posts
- 15. King of the Universe 1.861 posts
- 16. Vintage 29,3 B posts
- 17. F-35 24,7 B posts
- 18. Rachel Maddow 24,3 B posts
- 19. Anne 56,3 B posts
- 20. Christ Jesus 51,5 B posts
Who to follow
-
 Ullas Pedmale
Ullas Pedmale
@UllasPedmale -
 Chirag Jain
Chirag Jain
@chirgjain -
 Julia Kreiner
Julia Kreiner
@jmkreiner -
 MaizeZynski Hawaii
MaizeZynski Hawaii
@MaizeZynskiHI -
Jinliang Yang
@JinliangYang -
 Shujun Ou
Shujun Ou
@SigmaFacto -
 Tony Heitkam 🌱🔍💚 (left this site for 🟦🦋)
Tony Heitkam 🌱🔍💚 (left this site for 🟦🦋)
@toheitka -
 Salomé CARCY
Salomé CARCY
@CarcySalome -
 Dr Haifei Hu
Dr Haifei Hu
@lakeseafly1 -
 Keith Adams
Keith Adams
@KeithLAdams -
 Jeffrey Ross-Ibarra
Jeffrey Ross-Ibarra
@jrossibarra -
 Marcin Imieliński
Marcin Imieliński
@skimomiks -
 Aylwyn Scally
Aylwyn Scally
@aylwyn_scally -
 Yongfeng Zhou🍇🍷周永锋@[email protected]
Yongfeng Zhou🍇🍷周永锋@[email protected]
@yongfeng_zhou -
 Markus Stetter
Markus Stetter
@MGStetter
Something went wrong.
Something went wrong.