Zongming Ma
@ZongmingMaOccasionally a teacher, always a student. Data, model, method, theory. I enjoy all aspects of learning from data and experience.
Similar User

@RECOMBconf

@DengYanxiang

@jihk99

@shang_lulu

@MarcBosse5

@Pinkcigarette2

@wu_biostat

@GJCVeenstra

@zhizhid

@luoweicool

@CHO_HEMATO

@DongXu10710609

@sgqcheng

@EnCheng23

@mrjeffhsiao
Very happy to see the paper out today on NBT. See the thread by @NancyZh60672287 for some perspectives about batch integration that have motivated this work.
What gets erased when you integrate single cell data, and can you recover it? Finally, we know what happens and how to recover the lost signals. So excited to share CellANOVA, published online today in NBT! go.nature.com/4hZnzW5
The @Yale #HTAN Center, with a team led by @RongFan8, @Halene_lab, @ZongmingMa, & @MinaXu7, is constructing a comprehensive spatiotemporal molecular and cellular atlas of human lymphomas as a resource for mechanistic discovery. cancer.gov/about-nci/orga…

Extremely excited to join the @NCIHTAN consortium!!! Here is how we started our first Yale HTAN HuLymSTA center meeting 🥰 with @MinaXu7 @Halene_lab @ZongmingMa @YaleCancer @YaleEngineering @YaleMed

The @Yale #HTAN Center, with a team led by @RongFan8, @Halene_lab, @ZongmingMa, & @MinaXu7, is constructing a comprehensive spatiotemporal molecular and cellular atlas of human #lymphomas as a resource for mechanistic discovery. cancer.gov/about-nci/orga…

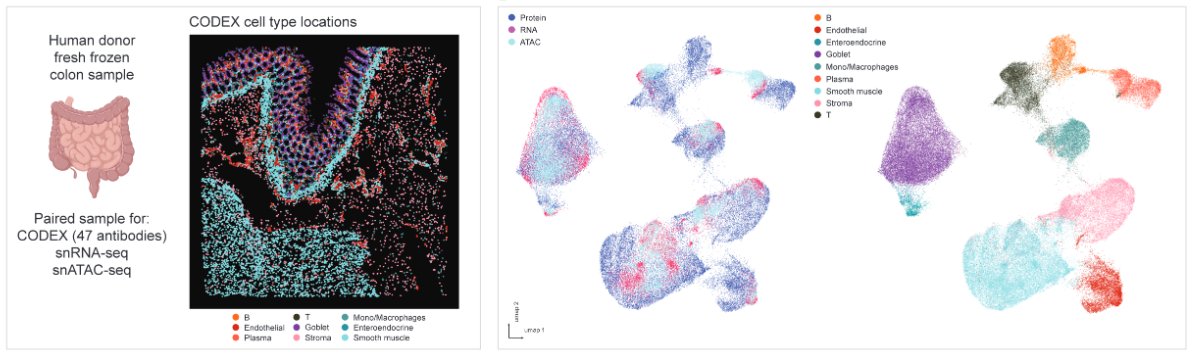
Excited to share our first preprint on a novel spatial multi-omics technology that co-profiles five modalities within the same tissue section. Great collab w/ @marekbartosovic and @DrMingyaoLi . Kudos to my postdoc Pengfei Guo and PhD student @LiranMao ! biorxiv.org/content/10.110…
Congratulations to @RongFan8, Mina L. Xu, MD, Stephanie Halene, MD, Dr Med, @HaleneLab and @ZongmingMa, who received an @NIH #U01 award to create the Center for Human #Lymphoma Spatiotemporal Atlas (HuLymSTA). yalecancercenter.org/news-article/y… @SmilowCancer @YaleMed @YNHH @yalepathology
It has been my great privilege to have had @nandysagnik as a student. May your career a wonderful journey! I am proud of you.

Congrats to @ZhuBokai and the wonderful team! Ever feeling frustrated with annotating cells from a spatial-omics dataset with a small targeted panel? CellSNAP helps by leveraging info from cell location and tissue image via a parallel GNN architecture. biorxiv.org/content/10.110…
1🔬/ Excited to share our latest preprint led by @ShuxiaoC , @Shenggao10 , @GarryPNolan , @SizunJ , @ZongmingMa , and with help from @shaleklab, Scott and more! biorxiv.org/content/10.110…

AI is all in the news, but scientists have been deploying its cousins machine learning and neural nets for nearly a decade in cancer research. One of the limitations in getting as many "eyes on the target" in studying cancer is that each KIND of target needs a different sensor…
Nobel Prize winner Katalin Karikó was 'demoted 4 times' at her old job. How she persisted: 'You have to focus on what's next' cnb.cx/3tqKkxm
Have you ever wondered if meaningful signals have been removed after batch correction? If so, please see below for a recent study with Jane Zhang, Nancy Zhang and other collaborators on how to retain meaningful signals and correct batch effects at the same time.
What gets erased when you integrate #singlecell data across samples/studies, and can you get it back? When samples are from e.g. healthy & disease, should you simply massage cells together? FINALLY, I feel we can answer this question: doi.org/10.1101/2023.0…
Cannot agree more! @GarryPNolan
There are hidden and embedded relationships in deep data that allow information transfer across knowledge domains (read that as RNA, protein, chromatic) even with "weakly linked" datasets. What does this mean? It means, to me, the end of worrying about running all 'omic…
Human languages have similar structures that describe the world. Researchers use the parallel languages of proteins, RNA, & epigenetics to describe cells/tissues. Such similarities can now fuse independent modalities w/ high fidelity. biorxiv.org/content/10.110…
United States Trends
- 1. Hunter 1,87 Mn posts
- 2. Cyber Monday 57 B posts
- 3. #IDontWantToOverreactBUT 1.031 posts
- 4. Enron 7.885 posts
- 5. #MondayMotivation 17,4 B posts
- 6. $CUTO 11,1 B posts
- 7. Yeontan 822 B posts
- 8. #GivingTuesday 4.018 posts
- 9. tannie 243 B posts
- 10. Burisma 67,6 B posts
- 11. Victory Monday 3.200 posts
- 12. #MondayVibes 4.865 posts
- 13. Good Monday 49,3 B posts
- 14. #Duolingo365 10,8 B posts
- 15. Scott Jennings 15,6 B posts
- 16. Pat Gelsinger 2.344 posts
- 17. Omer Neutra 12,8 B posts
- 18. DeFi 202 B posts
- 19. JUST ANNOUNCED 20,1 B posts
- 20. Fang Fang 11,9 B posts
Who to follow
-
 RECOMB Conference Series
RECOMB Conference Series
@RECOMBconf -
 Yanxiang Deng
Yanxiang Deng
@DengYanxiang -
 Hongkai Ji
Hongkai Ji
@jihk99 -
 Lulu Shang
Lulu Shang
@shang_lulu -
 Marc Bosse
Marc Bosse
@MarcBosse5 -
 Oliver Braubach
Oliver Braubach
@Pinkcigarette2 -
 Mike Wu
Mike Wu
@wu_biostat -
 Gert Jan C. Veenstra
Gert Jan C. Veenstra
@GJCVeenstra -
 Degui Zhi
Degui Zhi
@zhizhid -
 Wei Luo
Wei Luo
@luoweicool -
 Club CHO
Club CHO
@CHO_HEMATO -
 Dong Xu
Dong Xu
@DongXu10710609 -
 Qian Cheng
Qian Cheng
@sgqcheng -
 En Cheng
En Cheng
@EnCheng23 -
 Jeff Hsiao
Jeff Hsiao
@mrjeffhsiao
Something went wrong.
Something went wrong.