Stephen Rudd
@SAGRuddGenomics and bioinformatics. Impatiently looking for nirvana but discovering the world. Bioinformatics product management @nanopore; opinions & views my own
Similar User

@torstenseemann

@bffo

@strnr
@Awesomics

@druvus

@infoecho

@brent_p

@jameslz

@SEQanswers

@manuelcorpas

@BioInfo

@OmicsOmicsBlog

@johnquackenbush

@Alexbateman1

@mitenjain
GCI: A continuity inspector for complete genome assembly academic.oup.com/bioinformatics…

Very cool single-cell visualization package with support for TCR/BCR analysis and compatible with scRepertoire. Blown away by some of the viz ideas repo: github.com/pwwang/scplott… site: pwwang.github.io/scplotter/inde…

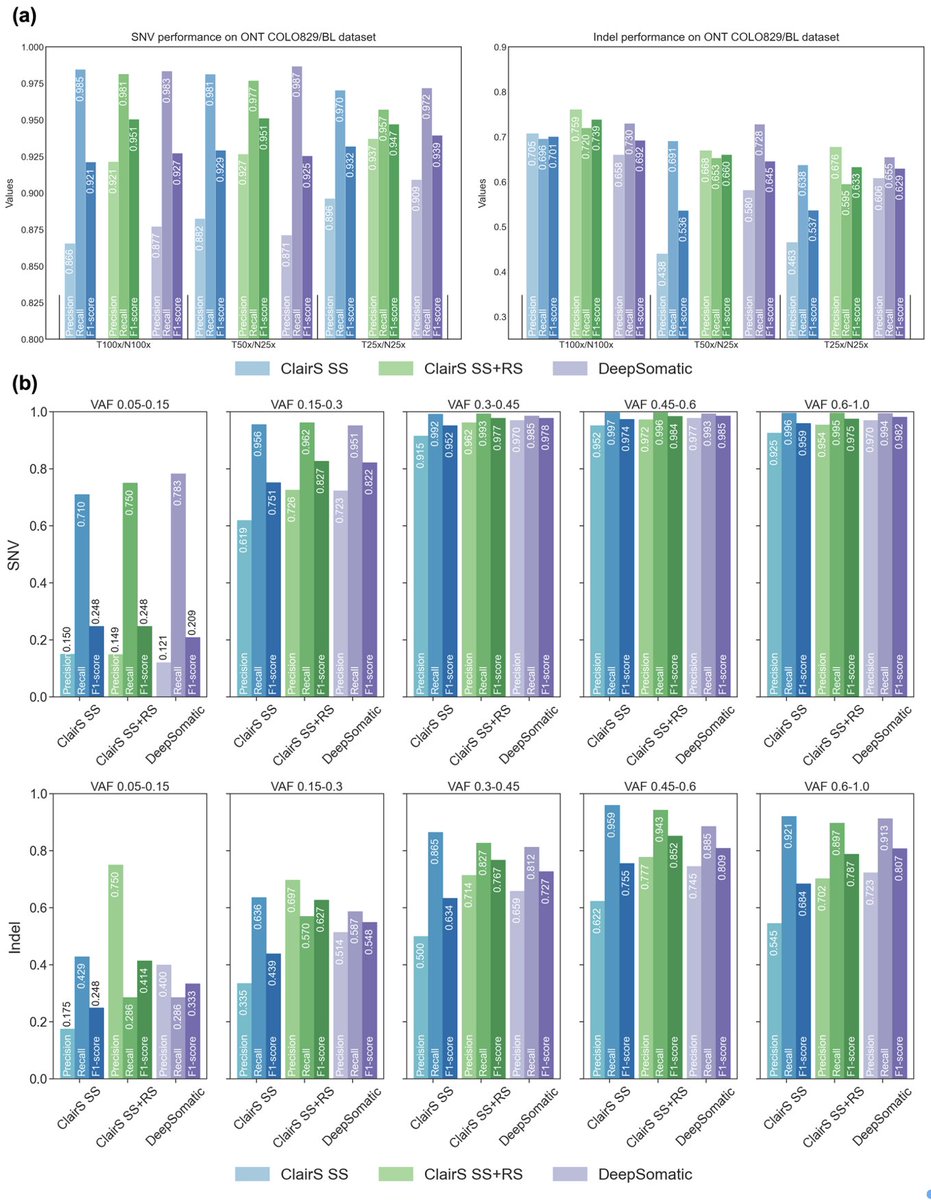
ClairS and ClairS-TO get a big performance bump. Check out the new versions and a technical note 🔗github.com/HKU-BAL/ClairS… to see how we have improved the performance of ClairS and ClairS-TO with new real cancer cell-line datasets and PoN. 1/

We continue to commit to a unified EPI2ME experience. We've launched the #EPI2ME cloud enabling EPI2ME in the cloud or locally - data analysis for all. #nanoporeconf
live from #NCMBOS2024 EPI2ME Cloud is finally released for everyone! we can’t wait to see what incredible bioinformatics analysis you all will do on the platform 🤘🚀

Learning German isn't about studying grammar or memorising Der, Die, Das. No, it's knowing 3 things: - when to interject with a solid "Wirklich!" - when to deliver a devastating "Doch!" - the optimum exasperated pitch for shouting "HALLO!" while in a supermarket queue.
A new dataset. Tumour/Normal pair on kit 14 5khz. Raw data and analysis available from our s3 bucket! labs.epi2me.io/colo-2024.03/
Lots of new workflow features and updates in this months @nanopore #EPI2ME release. Highlights include Salmonella Serotyping in wf-bacterial-genomes for all your food safety needs, and a new CNV caller in wf-human-variation (Spectre @sedlazeck) and… labs.epi2me.io/epi2me-24.03-0…
A key skill in bioinfo is assessment of raw seq data. Tools like IGV are useful to view aligned reads & variants. I've manually reviewed 1000s of variants and it's worth knowing what a real variant looks like! From Sirisha in the @nanopore #EPI2ME team: labs.epi2me.io/reviewing-bam
“Do you really need real-time?” - Natalia showing how ‘watchPath’ opens new horizons for at runtime data review #EPI2ME @IQCB_Mainz @nanopore @nextflowio

EPI2ME hackathon - @IQCB_Mainz @nanopore - Andrea deep diving into @nextflowio parallelisation and parameterisation; it can always be faster

The @nanopore EPI2ME team will be helping deliver this workshop. Come learn how we develop our workflows so you can deploy your own in EPI2ME!
Our advanced workshop for bioinformaticians offers hands-on experience in developing workflows for nanopore sequence analysis. Bring your laptop and skills and we will provide the coaching and challenges. Learn more: bit.ly/3HPYrQy #nanoporeconf #wymm

VCF files can be annoying to query if you don't know how. Andrea, who is developing our large human workflows which often produce unwieldy VCF files has written a blog post on how to do it: labs.epi2me.io/querying-vcf-f… @nanopore #EPI2ME
Absolutely mega release from the @nanopore EPI2ME team including big speed ups for wf-human-variation, a new BAC mode for wf-clone- validation, tumour only more for wf-somatic-variation and many many more improvements and bug fixes! labs.epi2me.io/epi2me-24.02-0… #AGBT #AGBT24
We get lots of questions on how input data should look for analysis with @nanopore EPI2ME workflows, Julian has put together a nice explanation! labs.epi2me.io/input-data/ If you have any requests for blog posts we'd be happy to hear your ideas!
We're making bioinformatics more accessible. With #EPI2ME anyone can now run any @nextflowio workflow — without any bioinformatics experience. Have a new workflow? Simply import it & share with your non-bioinformatician colleagues to use from the point & click interface. 1/2
You might have thought bed bugs was a very 2023 phenomenon.... wrong. Check out the new @nanopore tool "bed bugs" for the validation and generation of useful statistics from your bed files for adaptive sampling! tool: labs.epi2me.io/bed-bugs/ background: labs.epi2me.io/beg-bugs-intro/
EPI2ME: data analysis for all levels of expertise. Run preconfigured workflows from the EPI2ME application or the command line. Get comprehensive reports and rapid insights to make the most of your sequencing runs. Learn more: bit.ly/46BNZpV #AnythingAnyoneAnywhere
United States Trends
- 1. Bengals 38,3 B posts
- 2. Chargers 31,9 B posts
- 3. Herbert 22,3 B posts
- 4. Chiefs 145 B posts
- 5. Josh Allen 61,7 B posts
- 6. #BaddiesMidwest 11,9 B posts
- 7. Zac Taylor 2.418 posts
- 8. Burrow 2.182 posts
- 9. WWIII 131 B posts
- 10. 49ers 43,5 B posts
- 11. Russia 692 B posts
- 12. #RHOP 5.235 posts
- 13. Super Bowl 20,4 B posts
- 14. #CINvsLAC N/A
- 15. Jim Harbaugh 2.629 posts
- 16. Mahomes 40,1 B posts
- 17. #BoltUp 2.701 posts
- 18. Niners 8.670 posts
- 19. #DuneProphecy 2.371 posts
- 20. Geno 36,1 B posts
Who to follow
-
 Torsten Seemann
Torsten Seemann
@torstenseemann -
 BF Francis Ouellette
BF Francis Ouellette
@bffo -
 Stephen Turner 🦋 @stephenturner.us
Stephen Turner 🦋 @stephenturner.us
@strnr -
 Awesomics
Awesomics
@Awesomics -
 Andreas Sjödin
Andreas Sjödin
@druvus -
 Jason Chin
Jason Chin
@infoecho -
 brent pedersen
brent pedersen
@brent_p -
 Lei ZHANG
Lei ZHANG
@jameslz -
 SEQanswers
SEQanswers
@SEQanswers -
 Manuel Corpas
Manuel Corpas
@manuelcorpas -
 Justin H. Johnson
Justin H. Johnson
@BioInfo -
 Keith Robison
Keith Robison
@OmicsOmicsBlog -
 John Quackenbush
John Quackenbush
@johnquackenbush -
 Alex Bateman
Alex Bateman
@Alexbateman1 -
 Miten Jain
Miten Jain
@mitenjain
Something went wrong.
Something went wrong.