Stuart MacGowan
@stuartmac44Computational biologist | Protein structures & human variation | R & Python | Genetic effects on protein function | See: https://t.co/wzvNj7zDKI
Similar User

@gjbarton

@MarketaNovotna6

@tbrittoborges

@JavierUtges

@bioinformagic
@foreveremain

@sknepnek

@Marcus_Bage

@Jalview

@CCarvalheda

@PeterEGFIbrahim

@scientistsima

@MarekGierlinski

@biomadeira
We designed linear and cyclic peptide binders only from a protein target sequence. Different lengths; 46% had Kd in the micromolar range from a single sequence selection. biorxiv.org/content/10.110…
Zero-shot prediction of protein stability w/ inverse folding methods can be improved by re-running predictions on each residue in isolation, thereby identifying and removing the effect of local backbone geometry on prediction outcomes


Improving Inverse Folding models at Protein Stability Prediction without additional Training or Data biorxiv.org/cgi/content/sh… #biorxiv_biophys
The protein design & informatics group at @GSK is looking to hire a computational biologist in the UK and/or switzerland jobs.gsk.com/en-us/jobs/384…
We are seeking a great chemist to come and work with us and Bioascent as part of our new knowledge transfer partnership jobs.ac.uk/job/DHT622/kno… @UoDCeTPD @BioAscent #TPD
🥳As part of the organizing committee, I am excited to share the launch of the meeting Pandemic preparedness: Achievements, current challenges, and new frontiers, to be held in Trieste in November 2024. In collaboration with @alemarcelab @AreaSciencePark @ICGEB Please RT!
Developing innovative and effective responses to potential future pandemics: a topic to be discussed from 11 to 13 November in Trieste during the international #conference "Pandemic preparedness - Achievements, current challenges, and new frontiers" (#pnrr PRP@CERIC project)

To help scientists explore whether a genetic variant is likely pathogenic, and which regions of proteins are functionally important, we've integrated AlphaMissense data by @GoogleDeepMind into @ensembl, @uniprot & the AlphaFold database. ebi.ac.uk/about/news/tec…

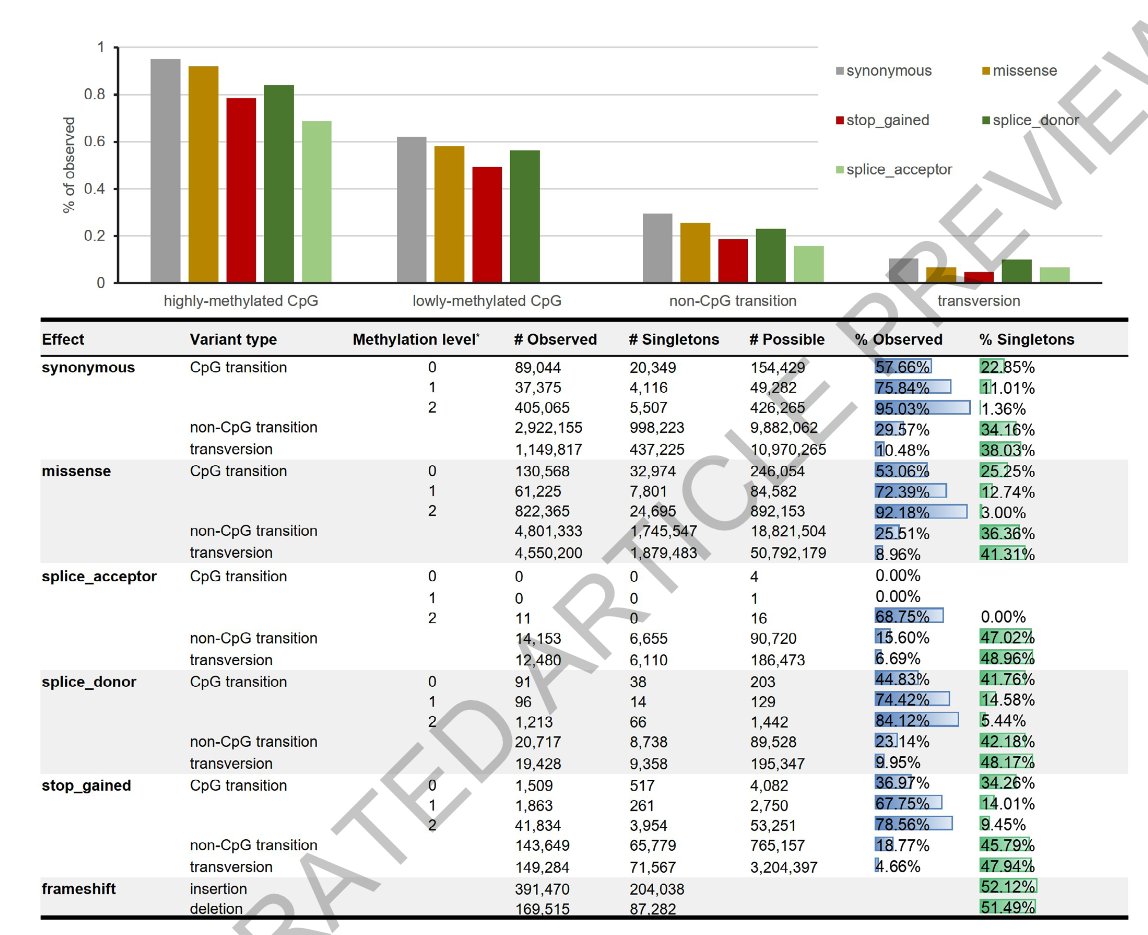
A deep catalogue of protein-coding variation in 983,578 (‼️) individuals nature.com/articles/s4158… 🧬🖥️ rdcu.be/dIweL


Wow this blows me away you can get roughly correct structure of pure electrolyte solutions out of this.


Thrilled to announce AlphaFold 3 which can predict the structures and interactions of nearly all of life’s molecules with state-of-the-art accuracy including proteins, DNA and RNA. Biology is a complex dynamical system so modeling interactions is crucial blog.google/technology/ai/…
(Editorial - this "let's own the functional consequences of all the mutations of all the bases in human" - in a combination of experimental and computational approaches I think is a super-sound foundational project we need - paging @mehurles and MAVE) #bog24
Knowing how to give a good talk is a crucial skill to have, but it's mostly not formally taught very well. Here's a 🧵on my strategy for how to prepare for the presentation, actually present it, and then answer the questions.
Ultra-fast and alignment-free variant effect prediction by training a small neural network on top of protein language model embeddings to predict the outputs from an MSA-based variant effect predictor. @LiniMuc Julius Schlensok @MarinaAbakarova @rostlab @LaineElodie @rostlab


NEW preprint: #PhyKIT is a command-line tool for analyzing multiple sequence alignments and phylogenies. Here, we detail #protocols for using PhyKIT to build #phylogenomic supermatrices, quantifying biases/#errors therein, identifying putative radiations & so much more.

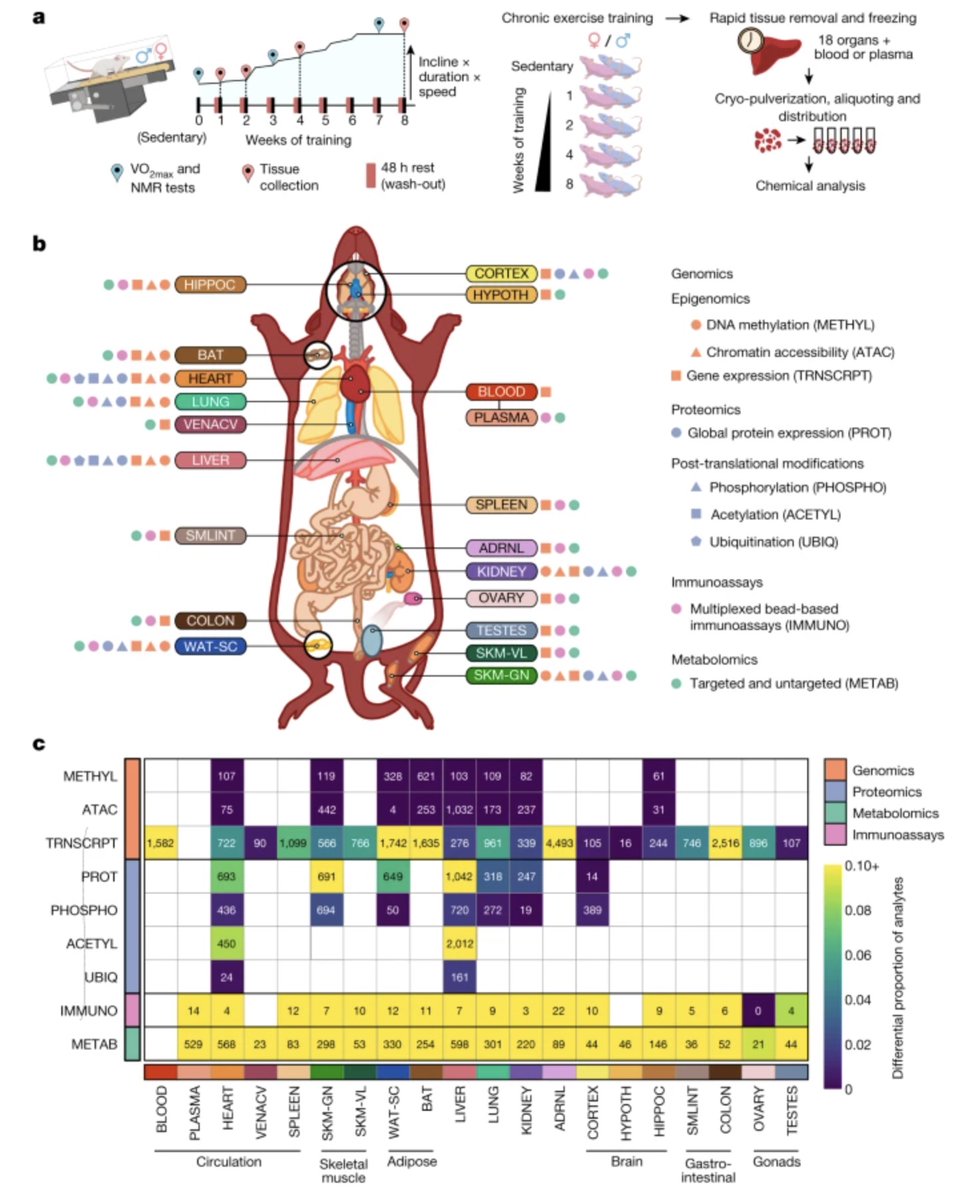
This is a very important and serious study - but - I can’t get the picture of rats running on tiny treadmills out of my head! Where is the emoji for this? I hope they were at least allowed some bangin’ beats 😁
Exercise may be the single most potent medical intervention ever known. Its benefits in prevention outstrip any known drugs: 50% reduction in the risk of cardiovascular disease, 50% reduction in the risk of many cancers, positive effects on mental health, pulmonary health, GI…

Highly variable NLRs have distinct genomic features, selective pressures, and mutational likelihood contributing to their super high amino acid diversity. But what's the mechanism??? How are these features established, and how do they change local mutation probability??
I have a seemingly simple question: in a GWAS, does including sex as a covariate alone enough to account for potential sex differences? What's the added benefit to doing a sex-stratified GWAS? Is there a paper on this? @sexchrlab @nmancuso_ @CharlestonCWKC might you know?
In two independent Science studies, researchers used time-resolved crystallography at an x-ray free electron laser to capture the process of #DNA repair from picoseconds to microseconds. Learn more on #DNADay ⬇️ 📄: scim.ag/6Jd 📄: scim.ag/6Jc
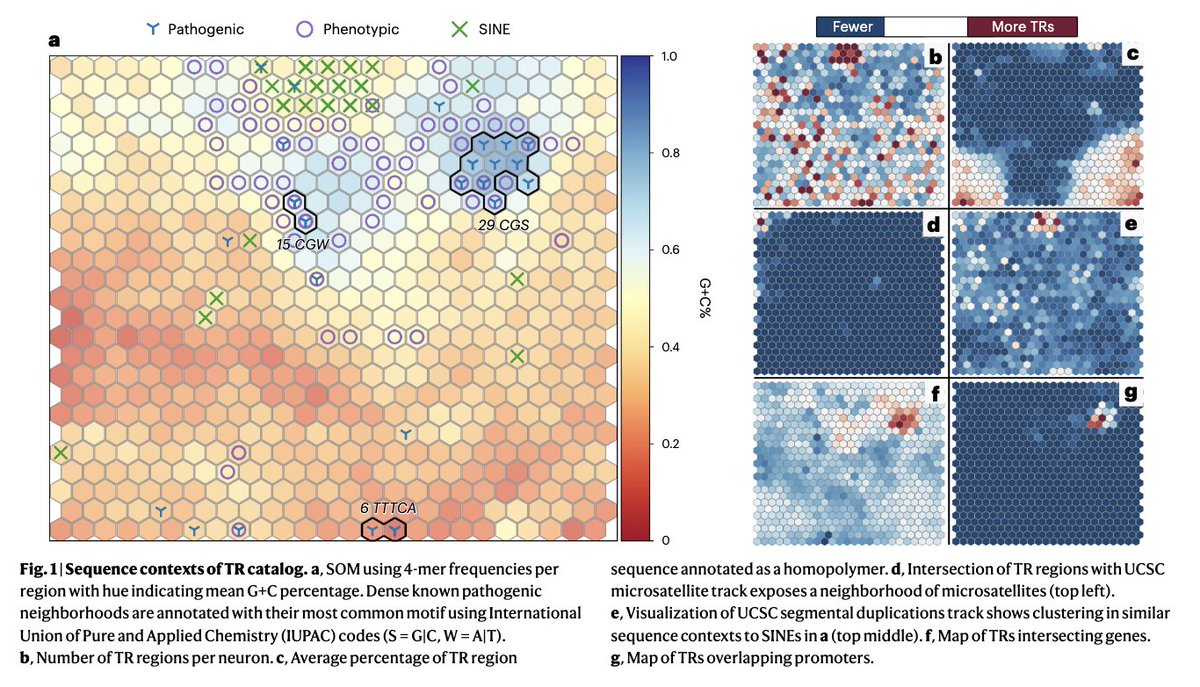
Tandem repeat @GenomeInABottle benchmark @NatureBiotech out today: rdcu.be/dFQNN . Characterization of 1.7 million TR + benchmark variants + new method to overcome var. representation issues! Great work lead by Adam E. @BCM_HGSC & great collab. from so many! (1/4)
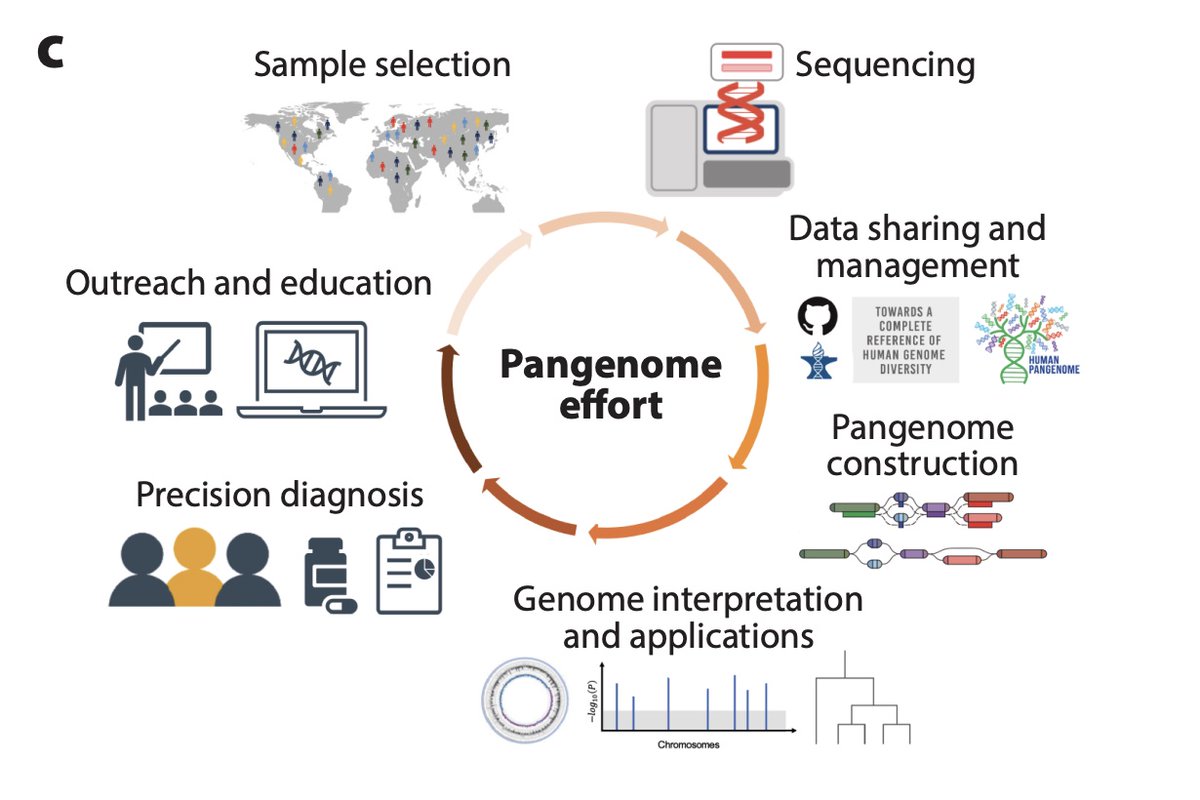
Excited to announce our new review: Beyond the Human Genome Project: The Age of Complete Human Genome Sequences and Pangenome References. w/ @BenedictPaten @rajivmccoy @khmiga @EimearEKenny et al. annualreviews.org/content/journa…
Introducing the Domainome... Site saturation mutagenesis of 500 human protein domains reveals the contribution of protein destabilization to genetic disease by @ToniBeltran13 @CRGenomica @sangerinstitute @BGI_Genomics biorxiv.org/content/10.110…
Happy to share the publication of our paper in @Nature Conformational ensembles of the human intrinsically disordered proteome Work led by @GiulioTesei and @AnnaIdaTrolle I'll post more later, but for now here is a link: nature.com/articles/s4158… and a short movie about the work
United States Trends
- 1. Serrano 176 B posts
- 2. #TysonPaul 82,5 B posts
- 3. #NetflixFight 43,3 B posts
- 4. #netflixcrash 6.720 posts
- 5. Shaq 9.822 posts
- 6. #buffering 7.233 posts
- 7. ROBBED 75,7 B posts
- 8. Ramos 64,3 B posts
- 9. Rosie Perez 8.751 posts
- 10. Cedric 15,6 B posts
- 11. My Netflix 52,4 B posts
- 12. Jerry Jones 7.205 posts
- 13. #boxing 36,1 B posts
- 14. Christmas Day 13,9 B posts
- 15. WTF Netflix 12,4 B posts
- 16. Love is Blind 4.433 posts
- 17. The Netflix 243 B posts
- 18. $NFLX 4.588 posts
- 19. Streameast 2.561 posts
- 20. Robbery 19,4 B posts
Who to follow
-
 Geoff Barton @gjbarton.bsky.social
Geoff Barton @gjbarton.bsky.social
@gjbarton -
 Marketa Novotna
Marketa Novotna
@MarketaNovotna6 -
 Thiago Britto-Borges
Thiago Britto-Borges
@tbrittoborges -
 Javier Sánchez Utgés
Javier Sánchez Utgés
@JavierUtges -
 Dr David Martin
Dr David Martin
@bioinformagic -
 Jim Procter
Jim Procter
@foreveremain -
 Rastko Sknepnek
Rastko Sknepnek
@sknepnek -
 Marcus Bage
Marcus Bage
@Marcus_Bage -
 Jalview
Jalview
@Jalview -
 Catarina A. Carvalheda Santos
Catarina A. Carvalheda Santos
@CCarvalheda -
 Peter E. G. F. Ibrahim
Peter E. G. F. Ibrahim
@PeterEGFIbrahim -
 Simone Altmann
Simone Altmann
@scientistsima -
 Marek Gierlinski
Marek Gierlinski
@MarekGierlinski -
 Fábio Madeira
Fábio Madeira
@biomadeira
Something went wrong.
Something went wrong.