David Schnettler
@JDSchnettlerFInactive. Find me on Bluesky or Mastodon https://t.co/986si2erZ8 https://t.co/OEJin8eGdc
Similar User

@PaolaLaurino

@CharlotteMiton

@hollfelderlab

@KlaraH_lab

@BirteHoecker

@SaacNicteh

@lacholt

@VijayJayaraman5

@21qiu

@d_zome

@erdogananisan

@SendkerFL

@AubelMargaux

@LasseMiddendorf
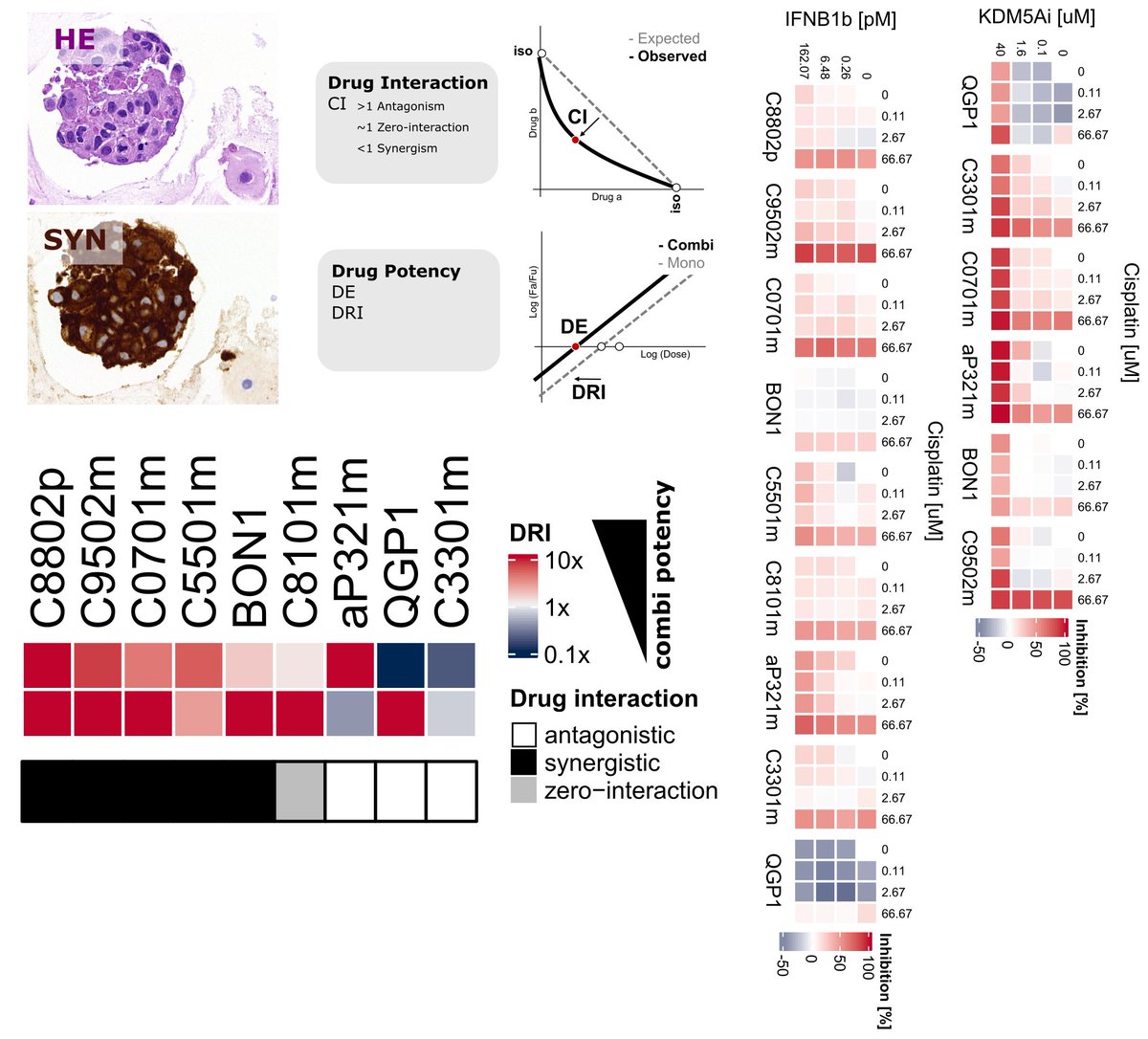
🚨𝐏𝐚𝐩𝐞𝐫𝐀𝐥𝐞𝐫𝐭🚨FreshOfPress 𝑶𝒖𝒓 #𝑵𝑬𝑻𝑮3𝑵𝑬𝑪 #𝒕𝒖𝒎𝒐𝒓𝒐𝒊𝒅 𝒔𝒄𝒓𝒆𝒆𝒏 @Nature_NPJ #LetsTalkAboutNETs #cancer #organoid ex vivo @CureNETs @Soc_Endo 𝐊𝐮𝐝𝐨𝐬 2col @unibern @ChariteBerlin @ng_tma @Bernomics @Swiss3RCC @MedFacultyUniBE doi.org/10.1038/s41698…
Über das "andauernde kulturoptimistische Vertrauen" und die vielfältige Geschichte der Migration in 🇩🇪: Starkes Plädoyer von @AgnesGehbald in der @faznet gegen die rechtsextreme Vereinnahmung des Begriffes #Remigration. ✊

yay, the new paper is out ✨ @MakMikhailCU and co playing with some odd lego bricks! read more on what are 25-mer peptides sprinkled with non-canonical AAs like and possible answers why we don’t find them in extant biology 🐍 @fried_lab @ELSI_Fujishima pubs.acs.org/doi/10.1021/ja…
Our latest preprint on how low temperature and nutrient scarcity lead to increased levels of stop codon miscoding in E.coli biorxiv.org/content/10.110…. Congratulations to all the authors! @RomeroRomeroML @JonasPoehls @mpicbg @csbdresden @LabShevchenko
Happy to see our paper from @hollfelderlab out in @NatureComms In this work we describe a #DirectedEvolution method for proteases based on protease inhibitors called alpha-2-macroglobulins (A2Ms). nature.com/articles/s4146…
Our collaboration w/ the @hollfelderlab, involving OPIG postdoc @mijr12, is now on bioRxiv. We benchmark a novel, readily-implementable flow platform to rapidly isolate antigen-specific antibody-secreting cells: biorxiv.org/content/10.110…
Tired of battling with noisy single-cell datasets? spinDrop to the rescue! We implement a sensitive droplet microfluidic system that discards empty droplets, cell multiplets and damaged cells from the sequenced dataset using droplet sorting. A 🧵(1/11) biorxiv.org/content/10.110…
Thrilled to see our parallel droplet generation and absorbance detection platform published in #an_chem - allowing the determination of enzyme kinetics in high throughput while minimising consumables and reagent consumption. @hollfelderlab @FGieLab doi.org/10.1021/acs.an…
Thomas Fryer, Joel Rogers et al. create a modular platform for protein engineering by ‘plug-and-play’ logic to achieve gigavalent display of functional proteins in 3D across a microbead, with control over valency and release. pubs.acs.org/doi/10.1021/ac…
Our new study came out few days ago in PNAS - Thankful to all members of the TAWFIK group and our amazing collaborators @SegevEinat lab and @LiamLongo lab for completing this work despite the loss Danny, our mentor and friend. I believe he would be proud. pnas.org/doi/epdf/10.10…
United States Trends
- 1. #SmackDown 36,4 B posts
- 2. Embiid 6.290 posts
- 3. #TheLastDriveIn 1.592 posts
- 4. #OPLive 1.541 posts
- 5. Standard 105 B posts
- 6. Paul George 2.214 posts
- 7. Melo 22,7 B posts
- 8. Daylight Savings Time 14,6 B posts
- 9. #SantaIsCoco N/A
- 10. #iubb N/A
- 11. Sixers 5.552 posts
- 12. Braun 8.043 posts
- 13. LA Knight 2.855 posts
- 14. Reggie Bush N/A
- 15. Michin 5.933 posts
- 16. OG Maco 4.273 posts
- 17. Mayorkas 16,9 B posts
- 18. Rare Exports N/A
- 19. Margaret Thatcher 2.617 posts
- 20. Sydney Sweeney 24,3 B posts
Who to follow
-
 Paola Laurino
Paola Laurino
@PaolaLaurino -
 Charlotte Miton
Charlotte Miton
@CharlotteMiton -
 Hollfelder Lab
Hollfelder Lab
@hollfelderlab -
 Klara Hlouchova lab
Klara Hlouchova lab
@KlaraH_lab -
 Birte Hoecker
Birte Hoecker
@BirteHoecker -
 Saacnicteh Toledo-Patino
Saacnicteh Toledo-Patino
@SaacNicteh -
 Lars Eicholt
Lars Eicholt
@lacholt -
 Vijay Jayaraman
Vijay Jayaraman
@VijayJayaraman5 -
 Kaiyu Qiu 邱锴宇
Kaiyu Qiu 邱锴宇
@21qiu -
 Dan Kozome
Dan Kozome
@d_zome -
 Ayşe Nisan Erdoğan
Ayşe Nisan Erdoğan
@erdogananisan -
 Franziska Sendker
Franziska Sendker
@SendkerFL -
 Margaux Aubel
Margaux Aubel
@AubelMargaux -
 Lasse Middendorf
Lasse Middendorf
@LasseMiddendorf
Something went wrong.
Something went wrong.