Deep Seq
@DeepSeqNottsCore Next Generation Sequencing facility at the University of Nottingham. Experts in Nanopore, Illumina and Sanger sequencing
Similar User

@The__Taybor

@mattloose

@AW_NGS

@Hasindu2008
@TJesse62

@fabian_theis

@mitenjain

@kirk3gaard

@johanneskoester

@anne_churchland

@cnag_eu

@ngsbioinfo

@martinalexsmith

@richard_compton

@edgenome
Hey @nanopore we have a weird blocking pattern on our flowcell... can you help?
We have an opportunity for someone to join our fabulous bioinformatics team - nottingham.ac.uk/jobs/currentva… - you will get to work with @nanopore @illumina @bionanogenomics @10xGenomics and more! Please retweet and dm for info!
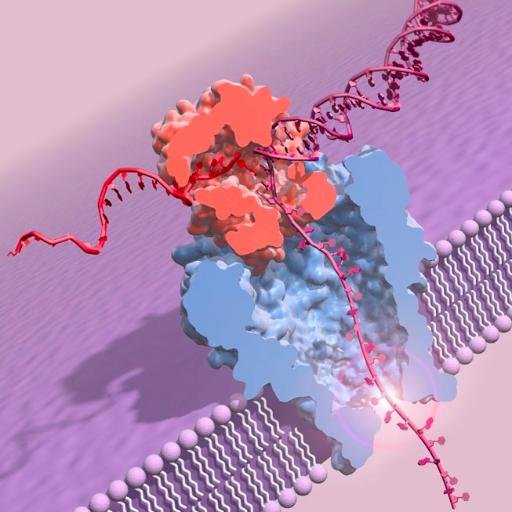
Rapid #genome-wide copy number profiling using adaptive sampling on @nanopore by @mattloose and the amazing team @DeepSeqNotts Real potential for clinical application.
This gif is from one sample of three run on a single @nanopore promethION flow cell. Frankly I'm super impressed at the performance though there is headroom for more improvements. We ran the same libraries as on gridION and have updated our preprint here. biorxiv.org/content/10.110…
This gif is from one sample of three run on a single @nanopore promethION flow cell. Frankly I'm super impressed at the performance though there is headroom for more improvements. We ran the same libraries as on gridION and have updated our preprint here. biorxiv.org/content/10.110…
Happy Christmas all!
We've put out a short preprint today showing some updates in Readfish for @nanopore adaptive sampling. biorxiv.org/content/10.110… @alexomics is giving a talk shortly at the #nanoporeconf about this - but a short thread here.
Earlier this week we put out a couple of preprints (doi.org/10.1101/2021.0… doi.org/10.1101/2021.0…) describing minoTour github.com/LooseLab/minot…, an @nanopore tool we have been developing for several years. A bit of a 🧵
I still remember the first time I saw data from @scalene showing ultra long reads (N50>=100kb) were possible. Since then we and others (especially @DrT1973) have worked to get longer reads. This weekend we tested the @circulomics @nanopore ultra long kits. Wow! 19.49 Gb N50 143kb

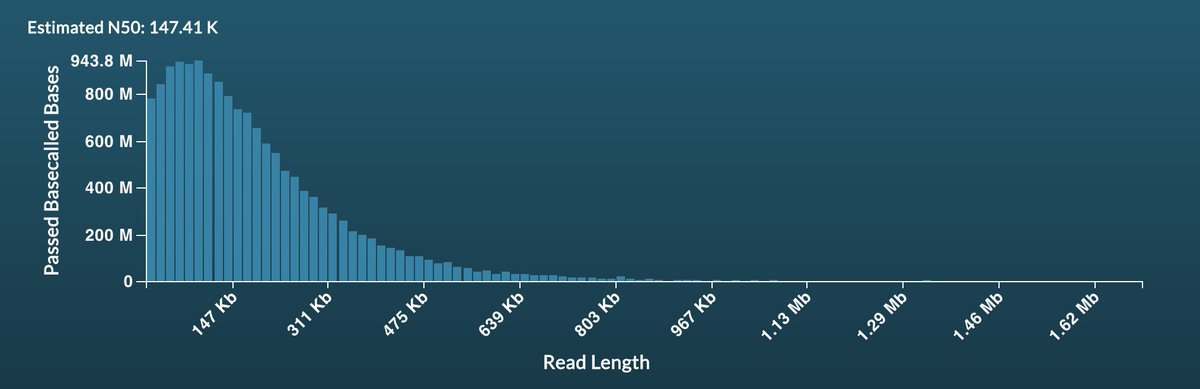
Well @nanopore - always seems to happen this time of year... bit of a bug in readfish but sure we will sort it out soon....
So - our ReadFish paper came out on Monday nature.com/articles/s4158… using GPU - but what if you only have a laptop? Here's an update from @alexomics to our tools that enable ReadFish on Mac, Linux or Windows with no additional GPU or compute. looselab.github.io/2020/12/02/rea… for details.

One for #nanoporeconf
So - our ReadFish paper came out on Monday nature.com/articles/s4158… using GPU - but what if you only have a laptop? Here's an update from @alexomics to our tools that enable ReadFish on Mac, Linux or Windows with no additional GPU or compute. looselab.github.io/2020/12/02/rea… for details.

Many application spin out of this on the fly selective sequencing. Tunable custom target panels to genome gap filling..... things get very interesting from here.
Great to see this paper out - loads of hard work from @alexomics and many others to get this completed - adaptive sampling on @nanopore sequencers targeting hundreds of genes to depths in excess of 40x on a single flowcell. nature.com/articles/s4158…
Great to see this paper out - loads of hard work from @alexomics and many others to get this completed - adaptive sampling on @nanopore sequencers targeting hundreds of genes to depths in excess of 40x on a single flowcell. nature.com/articles/s4158…
Announcing the Visium Spatial Gene Expression for FFPE Scientific Challenge! Share how you’d use Visium Spatial to unlock the full story of your FFPE tissue samples to win an experiment performed by us. Enter by 12/23. Learn more: bit.ly/2IIPjlk #spatial

About to start out 100th @nanopore run for covid samples with the team from @DeepSeqNotts . A sad milestone but amazing work by our local team including many from @TeamNUH and @PostdocPat all in support of the work of @CovidGenomicsUK .
Massive achievement for @TeamNUH Microbiology lab, one of the first labs in the UK to obtain UKAS accreditation for SARS-CoV-2 PCR. A true testament to the hard work & dedication of the micro team, maintaining <24-hr TAT & high quality, end to end service for our patients.




🧬 The total number of #COVID19 sequences now released is 16,380. The sequences and analysis are freely available to view 👉 cogconsortium.uk/data

Fantastic to be a part of this innovative distributed network.
Great to see this paper out - loads of hard work from @alexomics and many others to get this completed - adaptive sampling on @nanopore sequencers targeting hundreds of genes to depths in excess of 40x on a single flowcell. nature.com/articles/s4158…
United States Trends
- 1. Serrano 110 B posts
- 2. #TysonPaul 67,6 B posts
- 3. #NetflixFight 35,7 B posts
- 4. Ramos 62,9 B posts
- 5. #buffering 5.627 posts
- 6. #netflixcrash 2.943 posts
- 7. Rosie Perez 7.736 posts
- 8. My Netflix 45,4 B posts
- 9. ROBBED 48,7 B posts
- 10. Jerry Jones 6.569 posts
- 11. #SmackDown 65,9 B posts
- 12. WTF Netflix 10,6 B posts
- 13. Cedric 1.062 posts
- 14. Christmas Day 13,5 B posts
- 15. Love is Blind 4.181 posts
- 16. The Netflix 201 B posts
- 17. $NFLX 4.324 posts
- 18. Streameast 2.270 posts
- 19. Peter 150 B posts
- 20. Goyat 24,4 B posts
Who to follow
-
 Clive G. Brown
Clive G. Brown
@The__Taybor -
 Matt Loose
Matt Loose
@mattloose -
 Alexander Wittenberg
Alexander Wittenberg
@AW_NGS -
 Hasindu Gamaarachchi
Hasindu Gamaarachchi
@Hasindu2008 -
 Taco Jesse
Taco Jesse
@TJesse62 -
 Fabian Theis
Fabian Theis
@fabian_theis -
 Miten Jain
Miten Jain
@mitenjain -
 Rasmus Kirkegaard
Rasmus Kirkegaard
@kirk3gaard -
 Johannes Köster
Johannes Köster
@johanneskoester -
 Anne Churchland
Anne Churchland
@anne_churchland -
 CNAG
CNAG
@cnag_eu -
 NGS Bioinformatics
NGS Bioinformatics
@ngsbioinfo -
 Martin Smith
Martin Smith
@martinalexsmith -
 Rich Compton
Rich Compton
@richard_compton -
 Edinburgh Genomics
Edinburgh Genomics
@edgenome
Something went wrong.
Something went wrong.