Similar User

@_mixcr_

@ShikhaSinghzzz

@ynaazmi

@CamJohnWilson

@AnilBeh86779369

@soumendash07

@janduslab

@jeongwhan_choi

@gatechatl

@inmunoluna

@ChitraJoshi_93

@sisyga91

@keenious

@FrankCh72188182

@MurtadhaQ_Ali
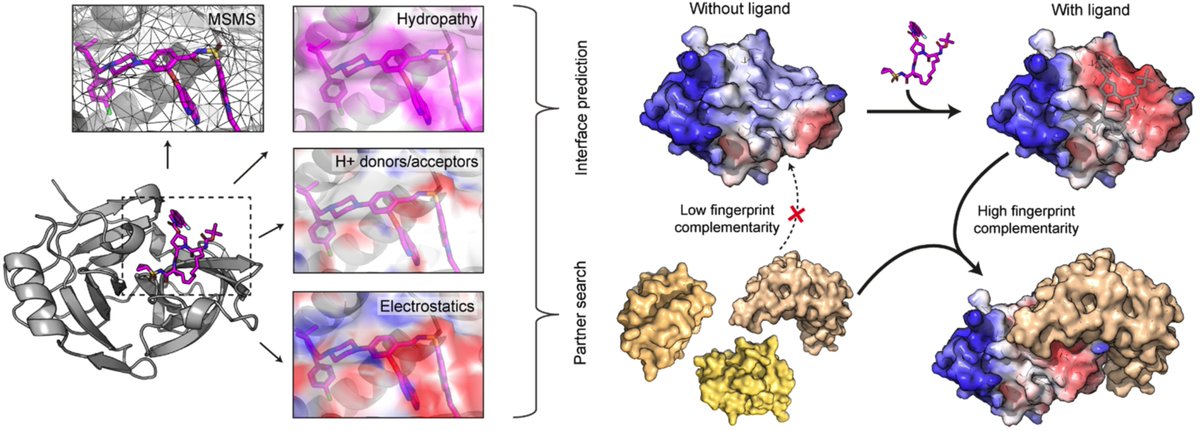
Happy to share a follow-up study is now published in Journal of Cheminformatics! Added more experiments and comparisons on the task of predicting protein-small molecule binding sites, an important step forward for drug discovery. Check it out! jcheminf.biomedcentral.com/articles/10.11…
How 3 Turing awardees republished key methods and ideas whose creators they failed to credit. More than a dozen concrete AI priority disputes under people.idsia.ch/~juergen/ai-pr…

🚀Excited to announce: Open-source AlphaFold3 implementation! 🚀 I am thrilled to announce one of the models we have been building for the last 8-weeks at Ligo - an open-source implementation of DeepMind’s frontier model, AlphaFold3! Here’s what we have learned, a thread (1/11):
To prepare a book chapter, I wrapped CLAPE as a Python package and will add more functions in the future. The book chapter will appear in Methods in Molecular Biology by the end of the year🥳 Thanks, Prof. Dukka from RIT, for the invitation. pypi.org/project/clape/
Generative AI, large language models, transformers, foundation models - what does it all mean and what is the potential impact on your research? Whether you're an AI novice or an AI expert, we invite you to read our Focus issue on advanced AI in biology. nature.com/nmeth/volumes/…
This is how machine learning engineers react after using GNNs by @Kumo_ai_team Thanks @TheMilennialDS for making this 😂 @PyG_Team
@jure i felt obligated to make this after reading your paper on GNN (Thank you Prof. Djordjevic for introducing Jure)
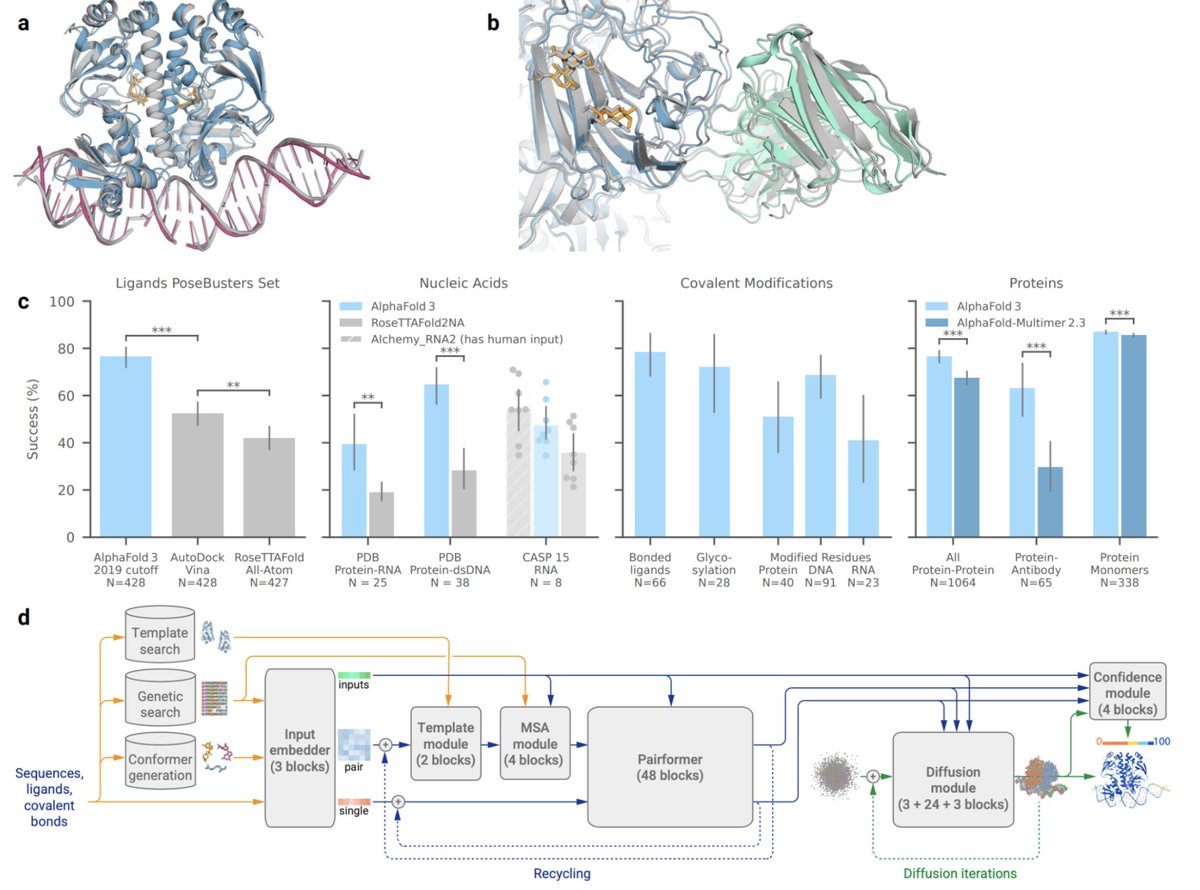
AlphaFold3 is insane. MutS protein (green) recognizes base mismatches in DNA (gray), after which it slides off to find MutL matchmaker protein (blue), which positions and activates MutH nuclease (pink) to nick a hemimethylated GATC sequence (red) to activate removal and repair of…
Super excited to be releasing AlphaFold 3 today, developed by @IsomorphicLabs and @GoogleDeepMind: our next generation AI model for predicting the biomolecular structures and interactions of proteins, DNA, RNA, small molecules, and more: bit.ly/44yfaCw 1/
Well deserved!
And... the first-ever ICLR test-of-time award goes to "Auto-Encoding Variational Bayes" by Kingma & Welling. Runner-up: "Intriguing properties of neural networks", Szegedy et al. The awards will be presented at 2:15pm tomorrow; there will be retrospective talks. Please attend!
Combining microdroplet screening with machine learning to improve enzyme kinetics. @MaxGantz_ @SimMat20 @pl219_Cambridge @FG_Mutti @hollfelderlab



Announcing NeurIPS Preschool Track This year, we invite preschoolers to submit machine learning research papers.

Fun adventure of our lab in small molecule design rdcu.be/dErPa pleasure to work with a bunch of cool people that made it possible @igashov @HannesStaerk @rneschneuing Clemence Vignac @vgsatorras @pafrossard @wellingmax @mmbronstein
everyone wants to train models no one wants to evaluate them
New preprint! In this project spearheaded by Anthony Marchand and @St_Buckley we designed new chemically-induced protein interactions and experimentally validated three binders targeting Bcl2:Venetoclax, DB3:Progesterone and PDF1:Actinonin complexes. biorxiv.org/content/10.110…
Glad to share that my first first-author paper has been published online in Briefings in Bioinformatics journal! More to come in my Ph.D. time! Protein–DNA binding sites prediction based on pre-trained protein language model and contrastive learning academic.oup.com/bib/article/25…
1/ Phew! Finally finished all 68 Python notebook exercises for "Understanding Deep Learning " book. Draft and notebooks at at: udlbook.com Pre-order your copy via: tinyurl.com/3yxs4hc6 Instructors can request an exam/desk copy via: tinyurl.com/mts4e3a6

Great work from my best friend Jiawei, and I know how much he endeavored to this paper. Congratulations and so proud of you!
Glad to share my PhD work published on @ScienceAdvances. We provide a systematic study on PAS domains: distribution, evolution, function, and roles as drug targets. Many thanks to my advisor @zhulinlab, with help of @VadimMGumerov. science.org/doi/10.1126/sc…
United States Trends
- 1. Cowboys 54,2 B posts
- 2. Cooper Rush 10,2 B posts
- 3. Texans 52,7 B posts
- 4. #WWERaw 58,5 B posts
- 5. Trey Lance 2.322 posts
- 6. Mike McCarthy 2.088 posts
- 7. Sixers 9.891 posts
- 8. Mixon 12,9 B posts
- 9. #HOUvsDAL 7.760 posts
- 10. Pulisic 19,3 B posts
- 11. Derek Barnett 1.472 posts
- 12. Aubrey 15,1 B posts
- 13. Pitre 1.231 posts
- 14. Zack Martin N/A
- 15. Keon Ellis N/A
- 16. #USMNT 3.277 posts
- 17. #AskShadow 6.124 posts
- 18. Turpin 3.108 posts
- 19. Guyton 1.186 posts
- 20. CJ Stroud 3.371 posts
Who to follow
-
 MiXCR
MiXCR
@_mixcr_ -
 Shikha Singh
Shikha Singh
@ShikhaSinghzzz -
 Liyana Azmi
Liyana Azmi
@ynaazmi -
 Cameron Wilson
Cameron Wilson
@CamJohnWilson -
 Anil Behera
Anil Behera
@AnilBeh86779369 -
 soumen dash
soumen dash
@soumendash07 -
 Janduslab
Janduslab
@janduslab -
 Jeongwhan Choi
Jeongwhan Choi
@jeongwhan_choi -
 Tim Shaw 🇺🇸..🇹🇼..🇺🇸
Tim Shaw 🇺🇸..🇹🇼..🇺🇸
@gatechatl -
 Inmunoluna
Inmunoluna
@inmunoluna -
 Chitra Joshi
Chitra Joshi
@ChitraJoshi_93 -
 Simon Syga
Simon Syga
@sisyga91 -
 Keenious
Keenious
@keenious -
 Frank Chen
Frank Chen
@FrankCh72188182 -
 Dr. Murtadha Ali
Dr. Murtadha Ali
@MurtadhaQ_Ali
Something went wrong.
Something went wrong.



















































































































