Richard Bethlehem
@rai_bethlehemAssistant prof. in NeuroInformatics & Director of Neuroimaging at the Autism Research Centre both at @Cambridge_Uni, fellow at @TrinCollCam
Similar User

@AndrewZalesky

@AFornito

@misicbata

@PTenigma

@bttyeo

@constable_todd

@MilhamMichael

@vdcalhoun

@amarquand

@ten_photos

@LindenParkes

@Nathan_Spreng

@petra_vertes

@LucinaUddin
Our latest global effort (led by twitterles Varun Warrier from @psychiatry_ucam and @CambPsych ) to map the genetic architecture of the human 🧠 is now live! Massive thank you to all co-authors!! nature.com/articles/s4158…
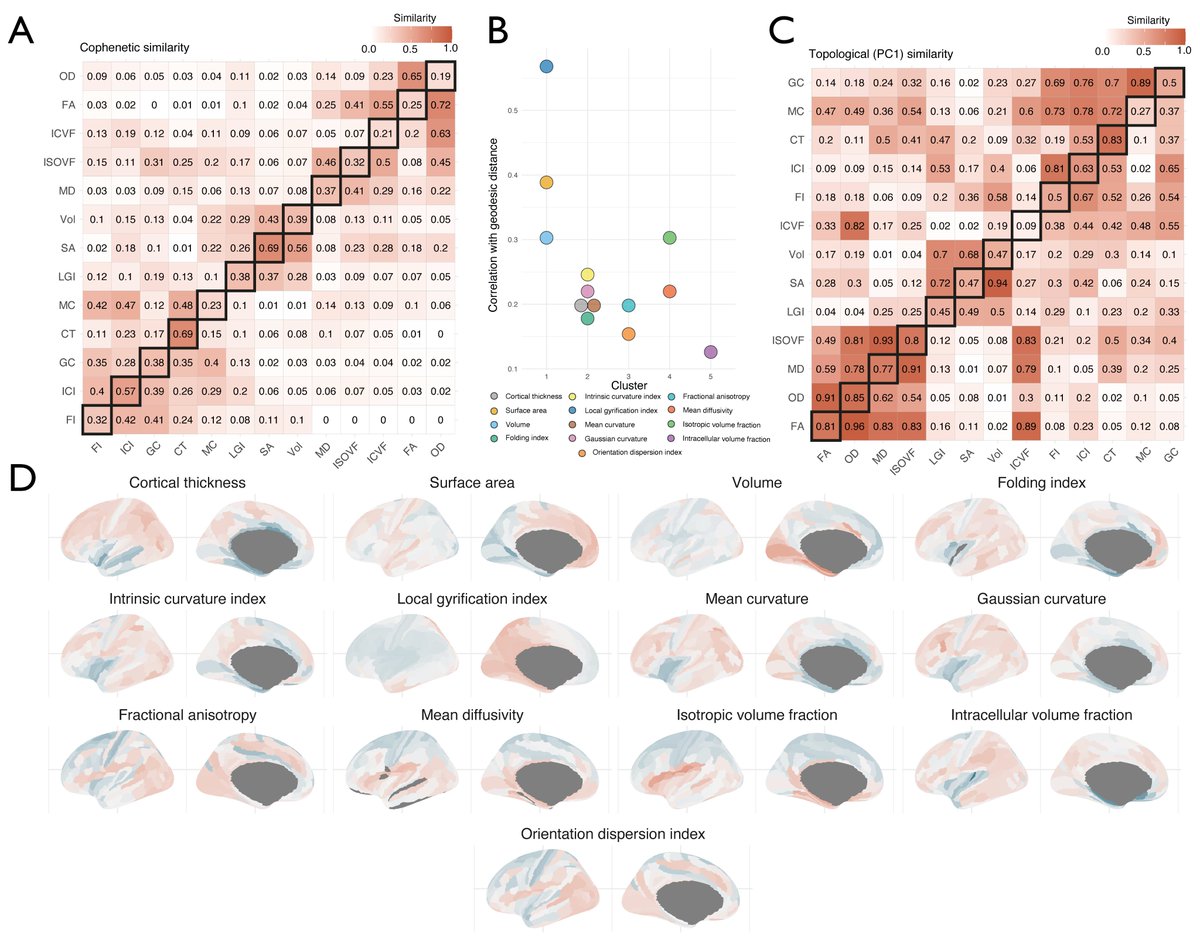
Very excited that our work has just been published in @NatureMedicine! nature.com/articles/s4159… A multimodal study reporting one-year cognitive, serum biomarker, and structural neuroimaging findings in 351 patients hospitalised with COVID-19🧠🦠📰 @gkwood @BenedictNeuro 1/9

#Covid cognitive deficits even 12-18months AFTER hospitalisation associated with reduced #brain volume on MRI and #brain-injury biomarkers in blood. OUT TODAY in @NatureMedicine
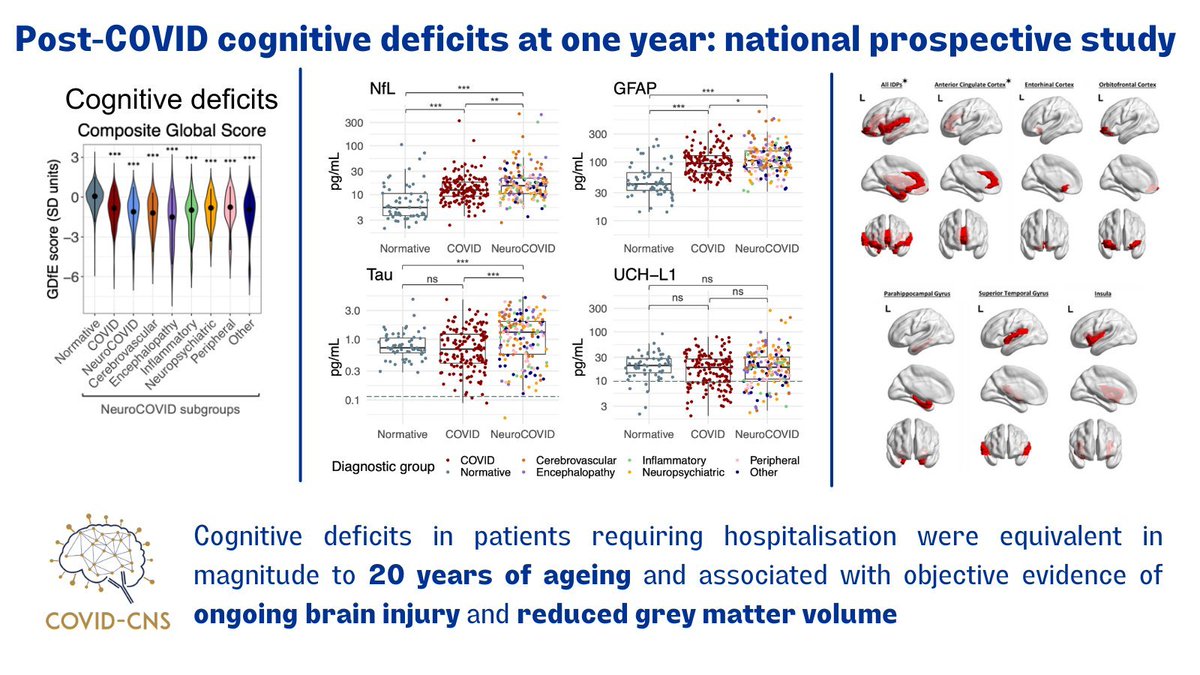
This was a huge group effort from people all over the country: @covidcns @Frosty_Immunity @hetheringtoncd @SarahAimeeB @Menon_Cambridge @Tim_R_Nicholson @rai_bethlehem @Cunningham_Colm @RunningMadProf @InfectNeuroLab A big shout out to the people who took part in the study!
Happy to share our new paper in @molpsychiatry We identified GMV reductions for different psychosis-related conditions based on centiles. An overlapping spatial distribution of neurobiological features was highly co-localized with the abnormal trajectories nature.com/articles/s4138…
Curious about human brain development through an animal model? 🧠🐒Check out our new preprint on Brain Charts of the Rhesus Macaque Lifespan! We've mapped normative trajectories for macaque brains using 1.5k MRI scans from PRIME-DE and collaborators! biorxiv.org/content/10.110…

Thrilled to share our preprint introducing ComBatLS, a new data harmonization method that preserves covariates’ (e.g. age and sex) effects on variance! (1/2) biorxiv.org/content/10.110…

Unlocking the full potential of your linear regression analysis starts with validating its suitability. Here’s a summary of how to validate assumptions before applying this model: 1️⃣ Linearity: The relationship between predictors (independent variables) and the response…

Idiocracy wasn't a comedy. It was a prophecy.
Registration page for the next Whistler Workshop on Brian Function - Feb 23-26, 2025 is open! Mark your calendars, reserve your spot! Program coming soon! RT! @sofievalk @bttyeo @AFornito @DrBreaky @LucinaUddin @Joana_Cabral__ @headspace_lab @markwoolrich medicine.yale.edu/mrrc/home/semi…
Does someone need to remind the president of the Spanish FA not to kiss any of the players if they win?
We shouldn't let this get swept away by the constantly changing news cycle. This is a premier data resource setting a dangerous precedent of pay to play. Not a policy conducive of a healthy scientific community.
Is anyone else annoyed that they've paid over £7,000 to access UK Biobank data, and now we have to also pay to analyse that data on the RAP?.....or am I just being a scrooge? I feel like being able to analyse the data *should* be included in the access fee?
The point is that computationally expensive analyses, for example requiring GPU processing, will likely be simply no longer affordable if we have to run all analyses on the cloud.
I did a quick estimation, and our connectome resource would have required up to 30,000 GBP to compute on RAP! Considering that similar computing resources are provided by research institutes worldwide, the centralized approach being enforced seems irrationally complicated!
Wow, with the new @uk_biobank rules, the dataset has just become way too expensive for the kind of computationally intensive work my lab does...
Really disappointing move by the @uk_biobank Worsening disparities in science and creating havoc for PhD students that have planned projects utilising UK biobank data. Are refunds available?
Wow, with the new @uk_biobank rules, the dataset has just become way too expensive for the kind of computationally intensive work my lab does...
I guess only companies will be able to afford to apply machine learning and AI to the @uk_biobank freezing out academic labs. Maybe that’s not the intent of this policy but that will be the consequence.
Wow, with the new @uk_biobank rules, the dataset has just become way too expensive for the kind of computationally intensive work my lab does...
The inability to download data and compute at local institutions will stifle innovation and crush the utility of the @uk_biobank The costs are prohibitively expensive for computationally intensive work and this will hinder most ML/AI/network-neuro research.
Wow, with the new @uk_biobank rules, the dataset has just become way too expensive for the kind of computationally intensive work my lab does...
Wow. I didn't read the email properly until now. Use cloud compute?? That's insane. The work we do leverages huge amounts of GPU compute from resources already bought and paid for. For sure just going to switch to ABCD
Wow, with the new @uk_biobank rules, the dataset has just become way too expensive for the kind of computationally intensive work my lab does...
Absolutely absurd considering the >10k GBP each research group has paid. Why should anyone feel ok to be forced to pay to use their cloud-based platform given the available cluster in our home Uni. This is a totally profit-driven decision and shame on uk_biobank.
Wow, with the new @uk_biobank rules, the dataset has just become way too expensive for the kind of computationally intensive work my lab does...
United States Trends
- 1. $PUMP 1.855 posts
- 2. Joe and Mika 2.285 posts
- 3. Good Monday 39,5 B posts
- 4. #MorningJoe N/A
- 5. #MondayMotivation 13,5 B posts
- 6. Mika and Joe 2.219 posts
- 7. Victory Monday N/A
- 8. #MondayVibes 2.237 posts
- 9. #18Nov 2.026 posts
- 10. Bengals 84,2 B posts
- 11. #MondayMood 1.636 posts
- 12. Chargers 72,8 B posts
- 13. 60 Minutes 40,7 B posts
- 14. WWIII 204 B posts
- 15. Rusia 128 B posts
- 16. Myron 13,4 B posts
- 17. Margaret Thatcher 5.837 posts
- 18. Herbert 36,7 B posts
- 19. $Marvin 7.737 posts
- 20. Ucrania 134 B posts
Who to follow
-
 Andrew Zalesky
Andrew Zalesky
@AndrewZalesky -
 Alex Fornito
Alex Fornito
@AFornito -
 Bratislav Misic
Bratislav Misic
@misicbata -
 Paul Thompson
Paul Thompson
@PTenigma -
 Thomas Yeo
Thomas Yeo
@bttyeo -
 Todd Constable (find me here @toddc.bsky.social)
Todd Constable (find me here @toddc.bsky.social)
@constable_todd -
 Michael Milham
Michael Milham
@MilhamMichael -
 Vince Calhoun @vcalhoun.bsky.social
Vince Calhoun @vcalhoun.bsky.social
@vdcalhoun -
 Andre Marquand
Andre Marquand
@amarquand -
 Thomas Nichols @[email protected]
Thomas Nichols @[email protected]
@ten_photos -
 Linden Parkes
Linden Parkes
@LindenParkes -
 Nathan Spreng
Nathan Spreng
@Nathan_Spreng -
 Petra Vertes
Petra Vertes
@petra_vertes -
 Lucina Uddin
Lucina Uddin
@LucinaUddin
Something went wrong.
Something went wrong.