Similar User

@GenomeInABottle

@Rachel_ONT

@tobiasmarschal

@Magdoll

@ethanmcqu

@catherinehgn

@ctsa11

@XLR

@mrvollger

@aquaskyline

@sergeynurk

@MedhatMahmoud_

@curious_coding

@csuhuangneng

@naechyun_chen
So happy to see this out today - our ultra-fast genome browser GW v1.0 #Genomics 🚀 biorxiv.org/content/10.110…

Very pleased to share our new online pre-print! We explore the spatial-temporal evolution of extrachromosomal DNA (ecDNA) in human glioblastoma. biorxiv.org/content/10.110…. A 🧵
Out today in Nature Methods, our first attempt make personalized pangenomes: rdcu.be/dTD3o In the future it will vital to subset pangenomes to just the relevant haplotypes for a compared sample. Work by @jltsiren, @27_matte, @par_esi, vg team, and many others.

🚨 Excited to share our latest paper in @CellCellPress identifying how under-replicated chromosomes in micronuclei become shattered by the Fanconi anemia pathway during mitosis. This work was led by @UTSWGradSchool student @JustinEngel17 🧵 (1/13): cell.com/cell/fulltext/…
🧬 Exciting new opportunities to join the AWMGS Bioinformatics team! We're looking for: 1️⃣ B6 Bioinformatician 2️⃣ B8A Lead Bioinformatician (Operational) Join us to help develop cutting-edge WGS, ctDNA and Somatic WGS pipelines and deliver new genomic tests for Wales.
A familial, telomere-to-telomere reference for human de novo mutation and recombination from a four-generation pedigree biorxiv.org/cgi/content/sh… #biorxiv_genomic
I’m very excited to share my PhD work on how age impacts tumor development and suppression. A huge thank you to @LabWinslow, @PetrovADmitri and all the co-authors for their support, wisdom, and patience (aging takes time!) over the past 5+ years. biorxiv.org/content/10.110… (1/n)
Sharing an update to 'sortedintersect' - a python library for fast interval queries. Can be used for searching a point or intervals against a reference set and is around 25% faster than other tested libraries: github.com/kcleal/sortedi…
Nanopore sequencing of 1000 Genomes Project samples to build a comprehensive catalog of human genetic variation medrxiv.org/content/10.110…
Here's a small package that integrates htslib with your Zig build, making it easier to work on genomics projects using #ZigLang. Check it out 👉 github.com/kcleal/zights #Genomics #Bioinformatics
Epigenetic clocks informed by Mendelian randomisation, nice idea! nature.com/articles/s4358…
🧬🎄Big update for GW (v0.9) brining loads of new features github.com/kcleal/gw/rele… 🔹 Desktop app with a new UI + more customisation 🔹 Even faster: 20 megabase regions rendered in ~1 sec 🔹 One-click BAM file access 👇 #Bioinformatics #Genomics
Exploring ~250,000 structural variants in GenomeSpy – just because we can! Ok, this is still work in progress and certain improvements are needed to make the visualization really useful. Stay tuned for updates! #biovis #bioinformatics #WebGL
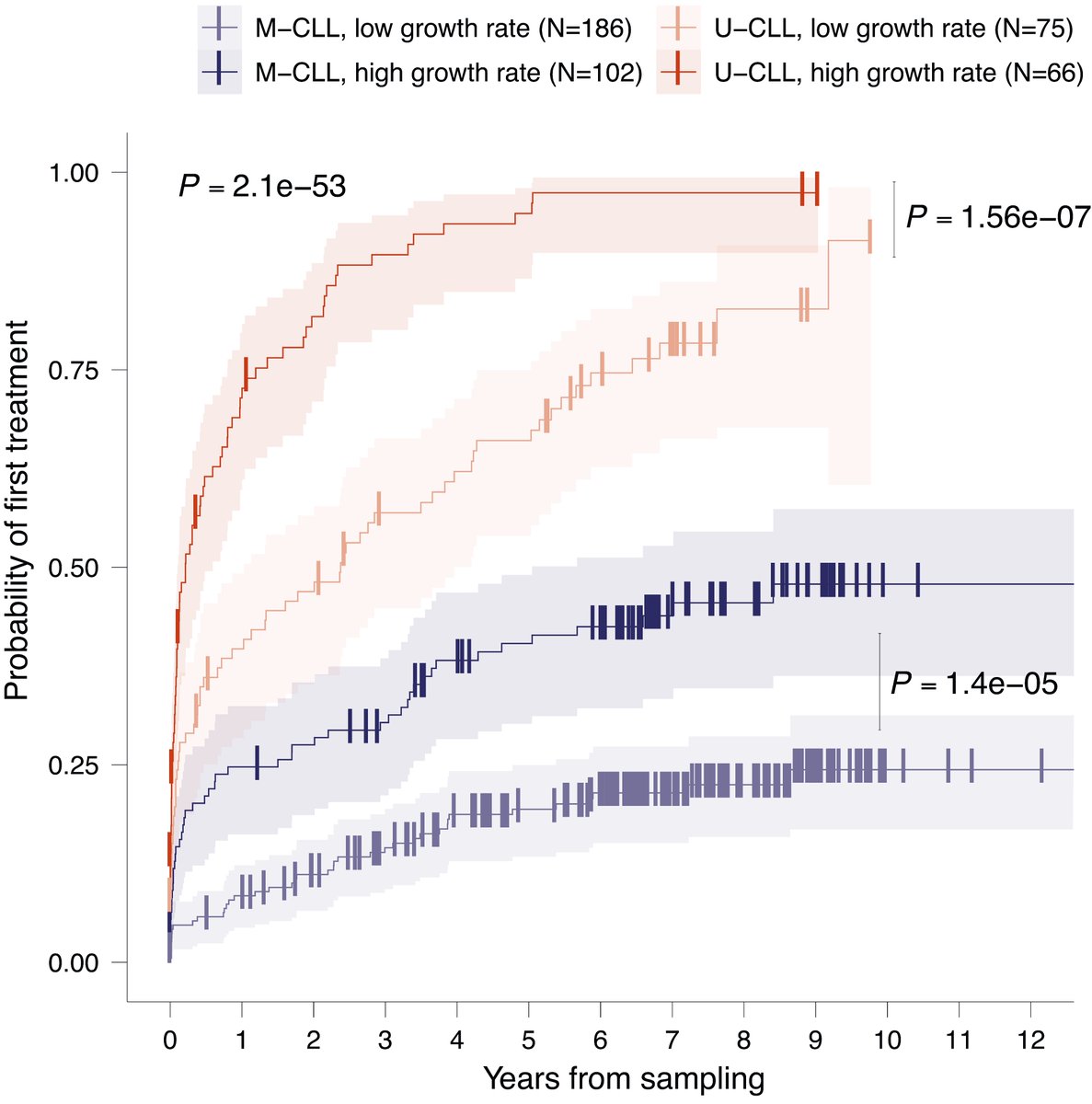
Does the evolutionary history of a cancer predict outcome? In our pre-print, we introduce a new method to measure a cancer’s past using bulk methylation data. In chronic lymphocytic leukaemia (CLL), these inferred histories are highly prognostic! (1/9) doi.org/10.1101/2023.1…

Amazed at how fast the new MacBook M2 Pro is. Called SVs on a 20X coverage Illumina sample using dysgu in only 7mins (2 threads) 🤯#Bioinformatics

Our CARD pilot paper with @KimberleyBill10 @cornelisblauw @BenedictPaten and many others is online! We establish a wet + dry lab protocol for single nanopore flow cell sequencing, which generates state-of-the-art SNP and SV calls + assemblies. rdcu.be/dl901
🧬Sharing a reproducible benchmark of SV callers using @PacBio HiFi data (Sequel II). Six runs were tested, only 8X coverage needed to detect over 90% of SVs! Boosting coverage to ~50X increased recall rates to 97.5% ⭐ #Genomics #Bioinformatics github.com/kcleal/SV_benc…

United States Trends
- 1. #chillguy 28,1 B posts
- 2. Good Sunday 57 B posts
- 3. #sundayvibes 5.602 posts
- 4. #SundayMorning 2.392 posts
- 5. Chuck Woolery 7.304 posts
- 6. #IPLAuction 93,9 B posts
- 7. #AskZB N/A
- 8. Bama 38,6 B posts
- 9. Lakers 41,7 B posts
- 10. Love Connection 5.428 posts
- 11. Auburn 36,1 B posts
- 12. Max Verstappen 140 B posts
- 13. F-35 21,3 B posts
- 14. Wheel of Fortune 3.763 posts
- 15. Pant 71,6 B posts
- 16. Lewis 121 B posts
- 17. Happy Birthday Anne N/A
- 18. Sainz 49,8 B posts
- 19. Ferrari 91,8 B posts
- 20. Oklahoma 50,7 B posts
Who to follow
-
 Genome in a Bottle
Genome in a Bottle
@GenomeInABottle -
 Rachel_ONT
Rachel_ONT
@Rachel_ONT -
 Tobias Marschall
Tobias Marschall
@tobiasmarschal -
 Liz Tseng
Liz Tseng
@Magdoll -
 Ethan McQuhae
Ethan McQuhae
@ethanmcqu -
 Catherine Hogan
Catherine Hogan
@catherinehgn -
 Chris Saunders
Chris Saunders
@ctsa11 -
 Armin Töpfer
Armin Töpfer
@XLR -
 Mitchell R. Vollger
Mitchell R. Vollger
@mrvollger -
 Ruibang Laurent Luo
Ruibang Laurent Luo
@aquaskyline -
 Sergey Nurk 🇺🇦
Sergey Nurk 🇺🇦
@sergeynurk -
 Medhat Mahmoud
Medhat Mahmoud
@MedhatMahmoud_ -
 Ragnar {Groot Koerkamp} 🦋
Ragnar {Groot Koerkamp} 🦋
@curious_coding -
 NENG HUANG
NENG HUANG
@csuhuangneng -
 Nae-Chyun Chen
Nae-Chyun Chen
@naechyun_chen
Something went wrong.
Something went wrong.