Ezzat El-Sherif
@ezzatelsherifBiologist working on the Systems Biology of Gene Regulation, Development and Evolution Assistant Professor, Department of Biology, UTRGV
Similar User

@PosnienLab

@michalis_averof

@KAPanfilio

@RebeizLab

@BertaVerd

@ChipmanLab

@MBattistara

@ecoevodevo_lab

@Schroeterfisch

@MarcHalfon
Out in @eLife! elifesciences.org/articles/84969 We use computational modelling, ATAC-seq and MS2 tagging in Tribolium to push for a model where the balance between 'dynamic' and 'static' enhancers enables the modulation of developmental timing and mediates dynamic gene expressions.
Curious about modeling gene regulatory mechanisms in embryonic pattern formation? My new manuscript guides you from the basics—starting with single-gene regulation modeled as chemical reactions—all the way to full GRNs. Easy-to-use MATLAB codes are included
From Genes to Patterns: A Framework for Modeling the Emergence of Embryonic Development from Transcriptional Regulation biorxiv.org/cgi/content/sh… #biorxiv_devbio
Arthropod Satellite meeting ready to rock! #EED2024 @ezzatelsherif

📢 Calling all Arthropod EvoDevo enthusiasts! Join us at the Arthropod Satellite Meeting during Euro Evo Devo 2024 in Helsinki @EED2024 Engage in talks and poster sessions. More info on registration and abstract submission here: shorturl.at/lnvP2 Pls retweet!

How does Pol II navigate the crowded, complex chromatin environment, surrounded by nucleosomes and other transcriptional activity? We are excited to share our preprint, a fantastic collaboration with @stergachislab, led by Tommy Tullius @mtcicero26 biorxiv.org/content/10.110… (1/5)
We don't know how the spatial patterns may have evolved from a temporal series (or vice versa). Wonderful work from @ezzatelsherif and others has proposed a wavefront-based model that could explain this. journals.plos.org/plosgenetics/a…
Konstantina's @KFilippopoulou Viewpoint was published today in the @FEBSJournal In it, we discuss instances of spatial and temporal patterning in insects and vertebrates towards understanding how patterning has evolved. febs.onlinelibrary.wiley.com/doi/abs/10.111…
I am having a great time #cshlmoet @CSHL discussing the mysteries of transcription. So many exciting science and fun discussions with Levine lab alumni (survivors)

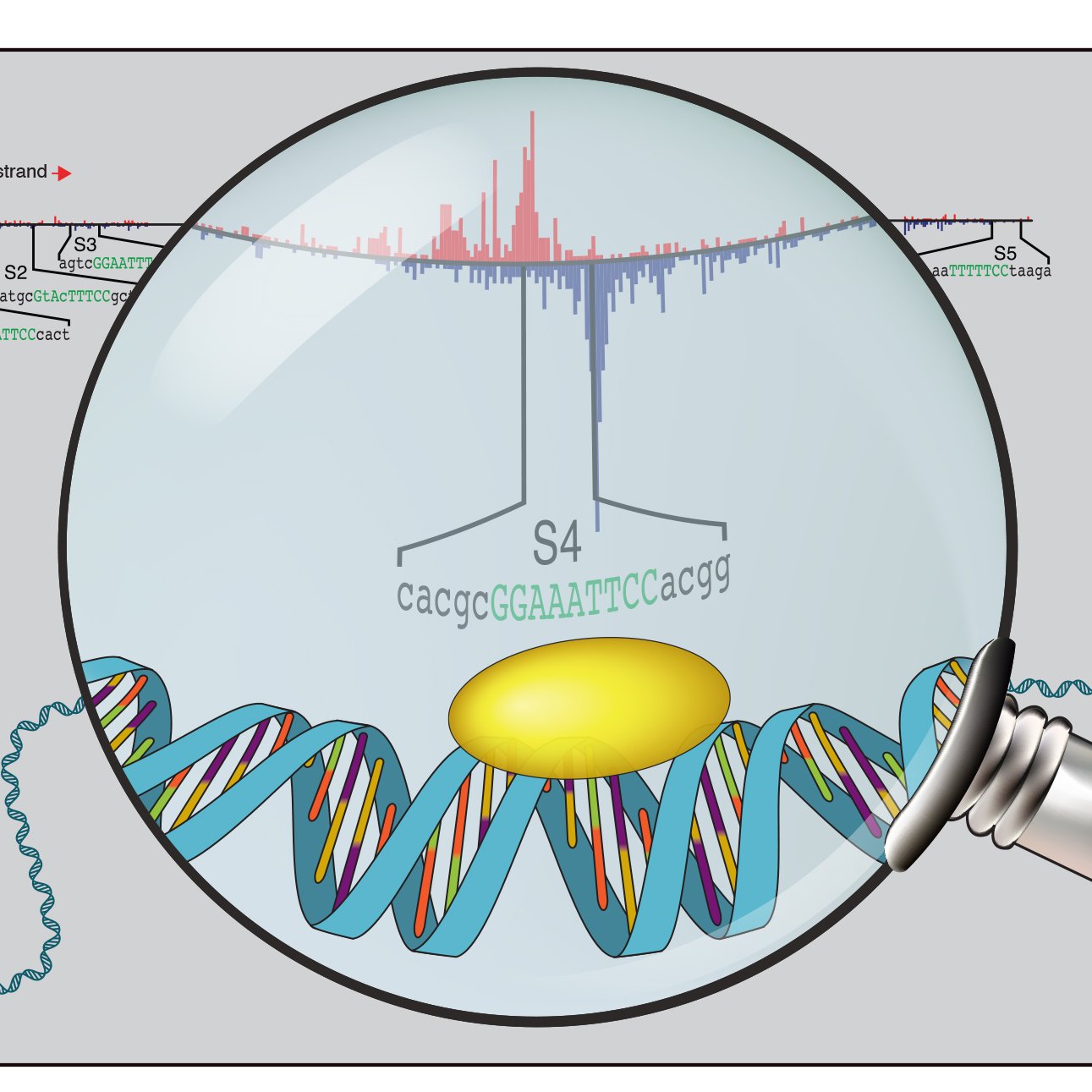
Our study on the cis-regulatory code combining deep learning and mechanistic experiments in the early Drosophila embryo is out! Very proud and grateful for everyone’s contribution 😊
Do only pioneer TFs open chromatin? No! Does motif affinity affect how well pioneering TFs work? Yes! Learn more by checking out our manuscript, now out in Developmental Cell! sciencedirect.com/science/articl…
Our latest: Cis-regulatory strategies for morphogen dependent cell fate decisions Two ways CREs interpret morphogens 1. Differential binding: CREs bind cell type TFs 2. Differential access: availability of CREs is cell type specific sciencedirect.com/science/articl…
#PhDposition studying #adaptation in European #Drosophila melanogaster populations. You will integrate #population #genomics data with genome wide expression & #chromatin accessibility data to unravel mechanisms underlying #natural #variation in quantitative phenotypic traits.
And finally, some skills that will be helpful: #bioinformatic and #statistical skills: population genetics/#genomics, quantitative genetics, #RNAseq, #ATACseq, integration of #multi_omics data; willingness to get involved in data generation (wet lab) is a plus.
United States Trends
- 1. $CUTO 8.030 posts
- 2. #JusticeforDogs N/A
- 3. ICBM 188 B posts
- 4. $EFR 2.121 posts
- 5. The ICC 254 B posts
- 6. Jussie Smollett 7.743 posts
- 7. Netanyahu 538 B posts
- 8. Denver 32,5 B posts
- 9. Illinois Supreme Court 7.768 posts
- 10. #KashOnly 41,4 B posts
- 11. Dearborn 6.702 posts
- 12. Volvo 5.927 posts
- 13. DeFi 122 B posts
- 14. chenle 128 B posts
- 15. #ATSD 10,6 B posts
- 16. #AcousticGuitarCollection 2.351 posts
- 17. Katie Couric 2.523 posts
- 18. #AtinySelcaDay 10,2 B posts
- 19. Flat 50,8 B posts
- 20. Bezos 39,8 B posts
Something went wrong.
Something went wrong.