Alex Rosenberg
@dna_rosenbergCEO and cofounder at @ParseBio. Making single cell sequencing easier and more scalable
Similar User

@WJGreenleaf

@JEFworks

@ParseBio

@slinnarsson

@JShendure

@hoheyn

@yimmieg

@morris_lab

@JD_Buenrostro

@dominic_grun

@arjunrajlab

@LGMartelotto

@KharchenkoLab

@vallens

@junyue_cao
This week, we launched Parse GigaLab, allowing researchers to scale single cell sequencing to billions of cells per year. 👉 Exponentially scalable 👉 Unmatched data quality 👉 Profile over 10 million cells or nuclei in a single run Start exploring: parse.bio/4fagAry
For those getting into single cell, this will be a great overview of experimental design and how to optimize sample prep. Led by our phenomenal Field Application Scientist Jorge Arturo Zepeda Martínez!
🎉Next week!🎉 Don't miss part two of our #SingleCell Educational Webinar series on Aug. 6th where we'll cover experimental design and sample prep for scRNA-seq experiments. Pacific Time Webinar: parse.bio/3ymylna Central European Time Webinar: parse.bio/46wzup2
Different webinars and resources from @ParseBio on how to get started.

Learn more about using Trailmaker in @ParseBio's webinar later this week with @kevinmbyrd: Mapping Interactomes: Precision Health through AI, Single-Cell, and Spatial Biology. Register below! resources.parsebiosciences.com/mapping-intera…
Single cell analysis can be hard. Fortunately, Trailmaker can help! Go from fastqs to publication ready figures all in the cloud. It clusters, auto-annotates cell types, computes differential gene expression, and more. And best of all you can use it for FREE!
This is a wild paper using @parsebio @mpsmela1 and his collaborators in the @geochurch lab figured out how to induce meiosis from human iPSCs. Very impressive and will have big impacts! biorxiv.org/content/10.110…
scRepertoire v2.0.3 now supports @ParseBio evercode TCR sequences. In coming changes, scRepertoire will also expand support for alternative python/R pipelines such as @KelvinTuong Dandelion workflow. Check it out at: borch.dev/uploads/screpe…

So excited to work with @ParseBio to share my work studying the role of Sox9 in promoting branching morphogenesis in mouse intrahepatic bile ducts (IHBDs). I will highlight how Parse scRNA-seq enabled discovery of a target to rescue IHBD branching in mice lacking Sox9.
Hannah Hrncir of @graczlab – Emory shares her recent work on the genetic controls of biliary duct formation. The pictures of these complex branching structures will amaze you, they’re beautiful. Virtual Live Webinar: May 1, 9:00 AM PST resources.parsebiosciences.com/sox9-links-bil…
See how Evercode WT v3 stacks up against Chromium Single Cell Gene Expression Flex Kit in mouse lymph node nuclei. Our latest tech note presents a head-to-head comparison of the two technologies. Explore the data 👉 parse.bio/4aRbzS5
Our next @cegs_ica Virtual Seminar (12PM on 4/16) @carol_dalgarno & Longda Jiang use Perturb-seq+@ParseBio+@UltimaGenomics to perturb signaling regulators across cell lines & infer causal regulatory changes from scRNA-seq/spatial data. Free registration: nygenome.zoom.us/meeting/regist…
Want to infer causal regulatory changes from scRNA-seq/spatial data? We combined Perturb-seq with @ParseBio + @UltimaGenomics to systematically perturb signaling regulators across six cell lines and learn their downstream targets. Preprint and data at biorxiv.org/cgi/content/sh…
And thanks a lot to @TreutleinLab @GrayCampLab for the supervision, @ETH_BSSE @IHB_Research @HFSP @EMBO for the support, and @ParseBio for the scalable scRNA-seq technology that makes this project possible
Congrats @HsiuChuan_Lin and team! Beautiful work!
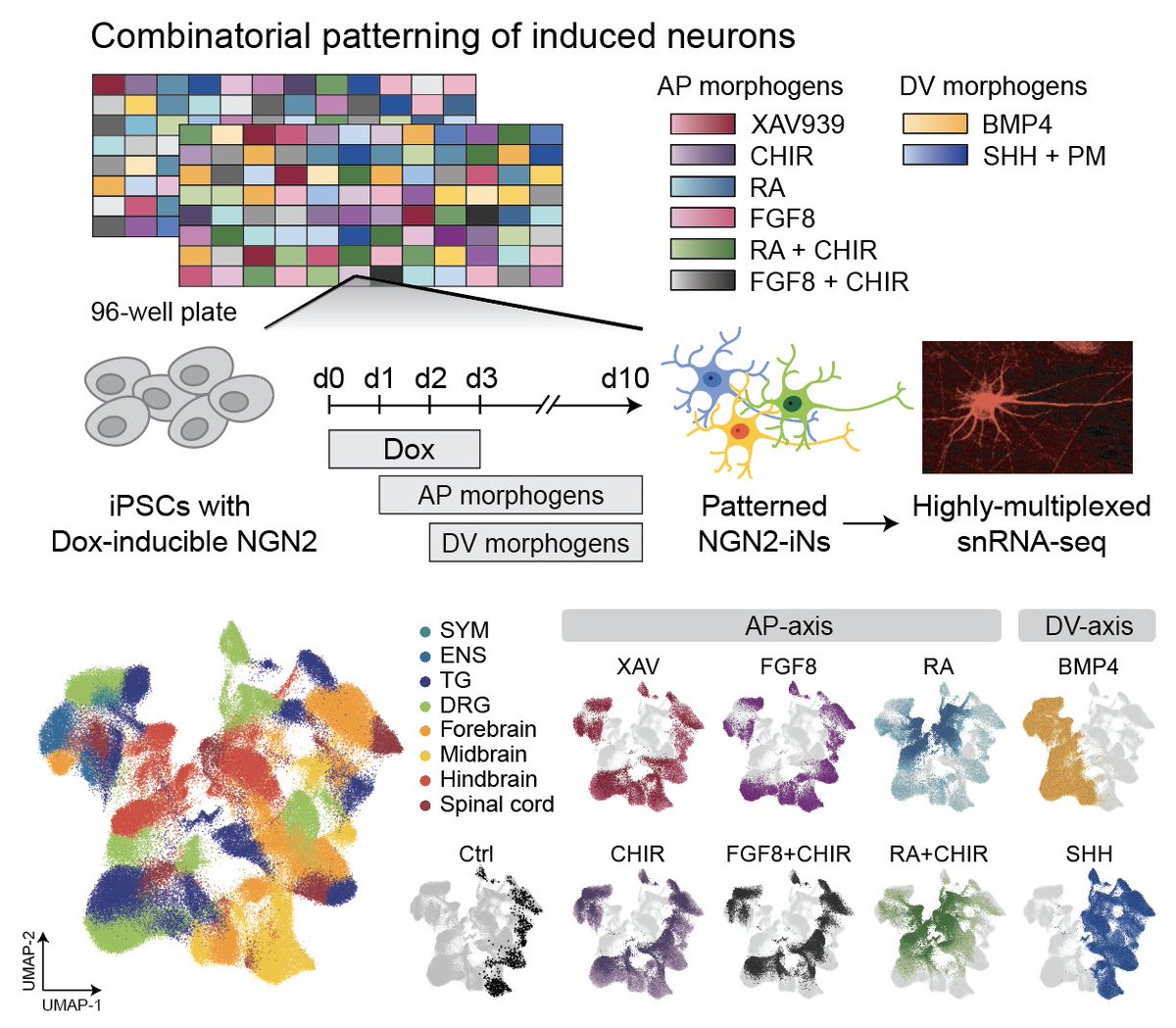
Programming diverse neuron subtypes from stem cells! Coupling patterning with proneural TF induction, we screened for 480 morphogen signaling modulations in ~700K cells. Great work with @JasperJanssens7 @TreutleinLab @GrayCampLab biorxiv.org/content/10.110…
Are you attending #ASHG23? Don't miss our featured education session with Rahul Satija (@satijalab) from the New York Genome Center. Lunch will be provided for the first 50 attendees.

Come chat with Efi (@epic_genetix) on November 2nd at #ASHG23 to learn about targeted single-cell transcriptome sequencing in 1 million healthy and leukemic bone marrow mononuclear cells.
"The data quality that we've got back with [Evercode] is really excellent and actually the best my lab has generated to date." - Prof. Dan Macqueen, Edinburgh University Learn more and explore the data from this experiment: parse.bio/46Lkl24
Starting my day off with some @ParseBio mini caffeine fixation kit! Wondering if I need to disclose this caffeine conflict of interest now…

United States Trends
- 1. Gaetz 767 B posts
- 2. Ken Paxton 9.624 posts
- 3. Attorney General 199 B posts
- 4. DeSantis 23,1 B posts
- 5. Volvo 15,1 B posts
- 6. Gary Gensler 27,1 B posts
- 7. Mike Davis 2.984 posts
- 8. Andrew Bailey 2.578 posts
- 9. 118th Congress 4.468 posts
- 10. Mark Levin 1.120 posts
- 11. Rubio's Senate 8.376 posts
- 12. ICBM 221 B posts
- 13. Dragon Believer 1.350 posts
- 14. Denver 41,9 B posts
- 15. The ICC 377 B posts
- 16. Netanyahu 740 B posts
- 17. Jussie Smollett 18,9 B posts
- 18. $SOLCAT 5.374 posts
- 19. Flat 55,5 B posts
- 20. Illinois Supreme Court 17 B posts
Who to follow
-
 William J. Greenleaf
William J. Greenleaf
@WJGreenleaf -
 Dr. Jean Fan
Dr. Jean Fan
@JEFworks -
 Parse Biosciences
Parse Biosciences
@ParseBio -
 Sten Linnarsson
Sten Linnarsson
@slinnarsson -
 Jay Shendure
Jay Shendure
@JShendure -
 Holger Heyn
Holger Heyn
@hoheyn -
 Jimmie Ye
Jimmie Ye
@yimmieg -
 Samantha Morris
Samantha Morris
@morris_lab -
 Jason Buenrostro
Jason Buenrostro
@JD_Buenrostro -
 Dominic Grün
Dominic Grün
@dominic_grun -
 Arjun Raj
Arjun Raj
@arjunrajlab -
 Luciano Martelotto 🛠🧬💻🇦🇺
Luciano Martelotto 🛠🧬💻🇦🇺
@LGMartelotto -
 PeterK
PeterK
@KharchenkoLab -
 Valentine Svensson
Valentine Svensson
@vallens -
 Junyue
Junyue
@junyue_cao
Something went wrong.
Something went wrong.