Alessandro Palma
@ale__palmaaPhD student at @HelmholtzMunich and @TU_Muenchen | ML and computational biology
Similar User

@AlieeHana

@LukasHeumos

@leon_het

@TillRichter6

@AnnaCSchaar

@p_tingying

@lauradmartens

@shitov_happens

@LucaEyring

@_sabrinarichter

@davidsebfischer

@Dominik1Klein

@g_palla1

@DrostFelix

@_yji_
1/6 Looking for neural estimators of entropic #OptimalTransport or simply cool applications of #FlowMatching? Excited by novel generative modeling tools for #SingleCell data? Check out our #GENOT #NeurIPS paper tinyurl.com/yc5deeke!

Deep learning with differential privacy can protect sensitive information of individuals. But what about groups of multiple users? We answer this question in our #NeurIPS2024 paper arxiv.org/abs/2403.04867 Joint work w/ @mihail_sto @ArthurK48147 @guennemann #Neurips (1/7)
Do you run functional assays? Wish you could get more results without having to scale? If you’re not using Prophet, you’re leaving potential on the table. (Warning: pitch not tweetorial)
We introduce Spatio-Spectral GNNs (S²GNNs) – an effective modeling paradigm via the synergy of spatially and spectrally parametrized graph conv. S²GNNs generalize the spatial + FFT conv. of State Space Models like H3/Hyena. Joint work w/ @ArthurK48147 @dan1elherbst @guennemann
Mixed Models with Multiple Instance Learning (MixMIL) received an Oral & Outstanding Student Paper award at @aistats_conf last week! 🏆 MixMIL enables accurate & interpretable patient label prediction from single-cell data by adding attention to GLMMs.#singlecell #MachineLearning


Dealing with undesired distribution shifts in unpaired translation tasks? Our #ICLR2024 paper shows how to mitigate them leveraging Unbalanced OT! We propose a method to incorporate unbalancedness into any neural Monge map estimator and demonstrate the benefits of unbalancedness.

Curious about how Self-Supervised Learning (SSL) is reshaping Single-Cell Genomics (SCG)? 🧬🤖 Our latest paper, "Delineating the Effective Use of Self-Supervised Learning in Single-Cell Genomics," offers an in-depth analysis. Thread [1/n] bioRxiv: doi.org/10.1101/2024.0…
I feel delighted about "Attention-based Multi-instance Mixed Models" being accepted to @aistats_conf as an oral! It is our attention-based MIL w/ @MaxIlse on steroids: with pre-trained embeddings, variational inference and SOTA incl. histopathology! Paper: arxiv.org/abs/2311.02455
I'm super excited to announce our new framework for exploratory electronic health record analysis "ehrapy". Although analysis is standardized for single-cell by seurat, bioconductor and scanpy, EHR analysis was until now the wild west. medrxiv.org/content/10.110…
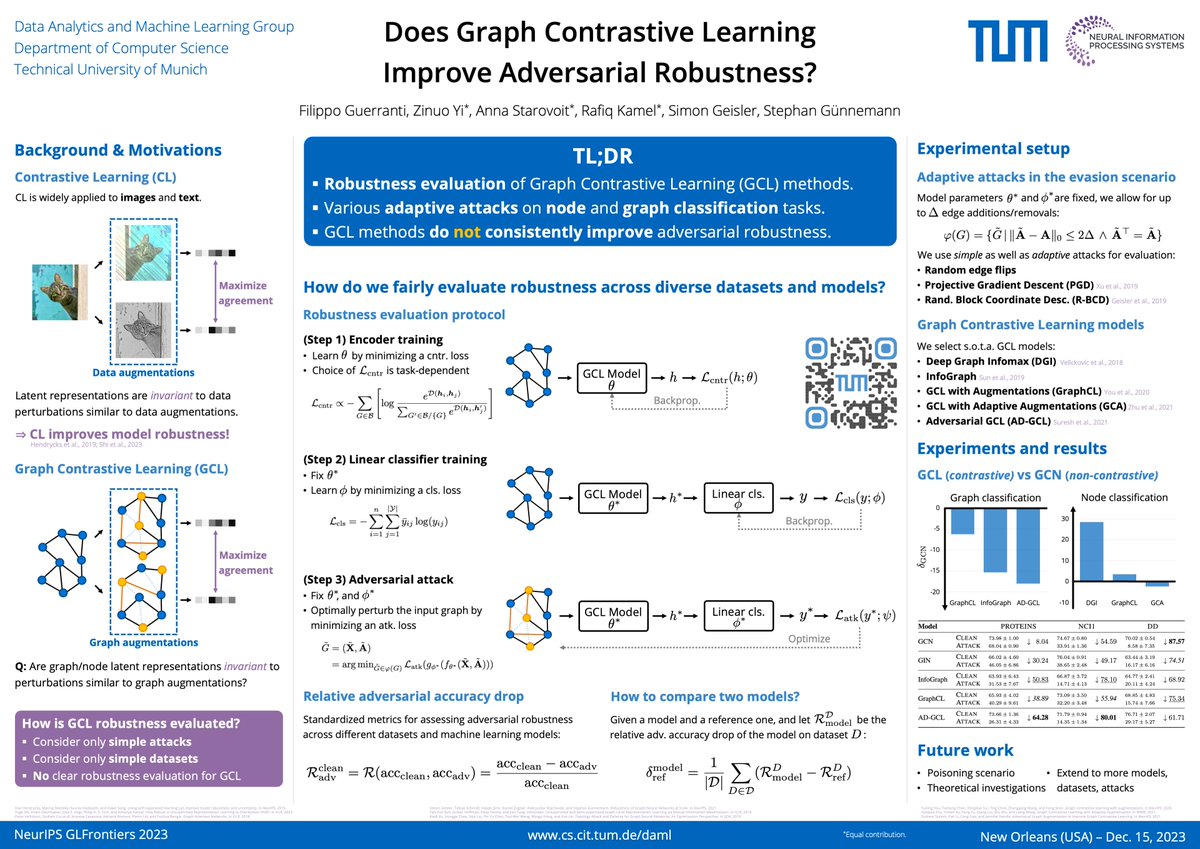
Does graph contrastive learning truly improve adversarial robustness? We answer this question in our work at #GLFrontiers @ #NeurIPS2023. Paper: arxiv.org/abs/2311.17853 Poster: 16 Dec 4:30pm Hall C2 Joint work with @ZinuoYi, @AnnaStrvt, @RafikiMazen, @geisler_si, @guennemann
Is binarization of scATAC-seq data really necessary? The conclusion from our analysis is that a quantitative treatment is in fact beneficial. Now out in Nature Methods! @gagneurlab @fabian_theis nature.com/articles/s4159… Many additions since the preprint 👇(1/n)

We will present our work “The geometry of hidden representations of large transformer models” at #NeurIPS2023 Great effort from @lucrevaleriani @DiegoDoimo #fracuturello @ansuin at @AreaSciencePark, and great collaboration with Ale Laio @SISSAschool! 📝 arxiv.org/pdf/2302.00294…

Want to learn cell state dynamics beyond RNA velocity? Led by @PhilippWeiler7 & @MariusLange8, and with @dana_peer, our new CellRank 2 enable fate mapping leveraging pseudotime, gene scores, time points or metabolic labeling in multi-view single-cell data. biorxiv.org/content/10.110…

1/10 image-based screening advanced drug discovery but scaling to massive perturbation space is hard! Given a cell image, we asked if we could predict the morphological effect induced by a perturbation! Led by @ale__palmaa & w @fabian_theis we propose IMPA tinyurl.com/yy4jfn4h
Introducing inVAE: Leveraging domain variability to learn conditionally invariant representations to unwanted biases. inVAE captures biological variations in single-cell datasets obtained from diverse conditions and labs. @HelmholtzMunich @KaplFer @soroorh1 @fabian_theis (1/7)🧵

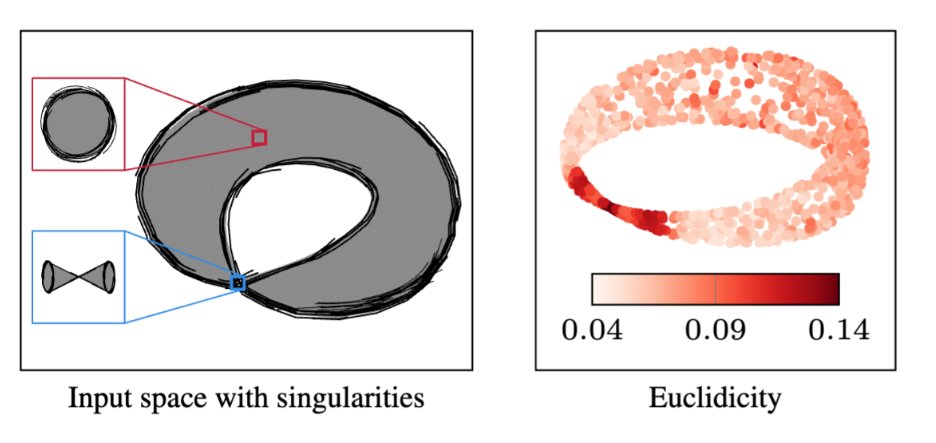
🎉Wow!🎉 Our paper 'Topological Singularity Detection At Multiple Scales' was accepted at #ICML2023. 👉We question the manifold hypothesis & develop a method to detect singularities, i.e. points that violate this 'manifoldness' assumption. #topology #MachineLearning 🧵1/n
Fresh on bioRxiv: @sophiaMaedler and I developed ✨SPARCS✨, a technology for microscopy-based genome-wide genetic screens in human cells🔬🧬 biorxiv.org/content/10.110… This work was enabled by the amazing collaborative environment created by @hornung_lab and @labs_mann 🧵👇
1/10 Looking for a tool to map cells across time and space? We introduce moscot-tools.org, a scalable framework for #optimaltransport (#OT) applications in single-cell genomics! bit.ly/3MiICFc

United States Trends
- 1. $CATEX N/A
- 2. $CUTO 7.470 posts
- 3. #collegegameday 2.004 posts
- 4. $XDC 1.365 posts
- 5. DeFi 105 B posts
- 6. #Caturday 7.673 posts
- 7. Henry Silver N/A
- 8. #saturdaymorning 3.066 posts
- 9. Jayce 80,4 B posts
- 10. Good Saturday 36,1 B posts
- 11. #MSIxSTALKER2 5.886 posts
- 12. #Arcane 297 B posts
- 13. Senior Day 2.883 posts
- 14. Pence 84,8 B posts
- 15. Renji 3.551 posts
- 16. Fritz 8.405 posts
- 17. Fishers N/A
- 18. Zverev 6.743 posts
- 19. McCormick-Casey 28 B posts
- 20. Tyquan Thornton N/A
Who to follow
-
 Hana Aliee
Hana Aliee
@AlieeHana -
 Lukas Heumos
Lukas Heumos
@LukasHeumos -
 Leon Hetzel
Leon Hetzel
@leon_het -
 Till Richter
Till Richter
@TillRichter6 -
 Anna Schaar
Anna Schaar
@AnnaCSchaar -
 Tingying Peng
Tingying Peng
@p_tingying -
 Laura Martens
Laura Martens
@lauradmartens -
 Vladimir Shitov
Vladimir Shitov
@shitov_happens -
 Luca Eyring
Luca Eyring
@LucaEyring -
 Sabrina Richter
Sabrina Richter
@_sabrinarichter -
 David Fischer
David Fischer
@davidsebfischer -
 Dominik Klein
Dominik Klein
@Dominik1Klein -
 Giovanni Palla
Giovanni Palla
@g_palla1 -
 Felix Drost
Felix Drost
@DrostFelix -
 Yuge Ji
Yuge Ji
@_yji_
Something went wrong.
Something went wrong.