Xiuqi Chen
@XiuqiChenPostdoctoral researcher @tudelft Trying to sequence proteins with nanopores
Similar User

@d1kozan

@_chrisT3na

@simonational

@smyan__

@p_deford

@KevinRhine_

@supmerlin

@MoutonTaylar

@tehhoyen

@MRStarostik

@taschroer

@maryamyamadi

@radhikajangi

@CaiPoz
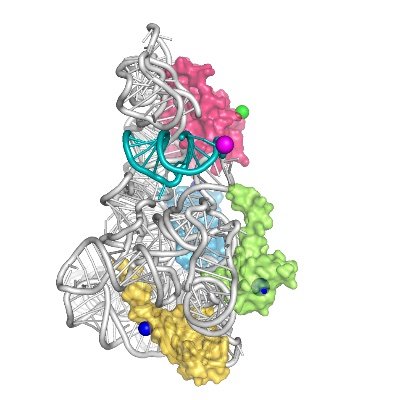
Very happy to share our latest work @acsnano on applying #nanopore to protein analysis! With the motor protein Hel308, we translocate DNA-peptide conjugate through MspA and successfully distinguish sulfation and phosphorylation at #singlemolecule level.

Nature research paper: Tracking transcription–translation coupling in real time go.nature.com/3VLPiA7
Could one envision a synthetic receptor technology that is fully programmable, able to detect diverse extracellular antigens – both soluble and cell-attached – and convert that recognition into a wide range of intracellular responses, from transgene expression and real-time…

I am really excited to announce the first Hubrecht Symposium on March 13th 2025! We will organize these yearly events on a specific topic in molecular & developmental biology to emphasize the importance of basic research for Dutch science. Free of charge! hubrecht.eu/hubrecht-sympo…

BLUESKY IS THE NEW TWITTER! Please join me there at bsky.app (or use that handy 'tweedeck-like' app deck.blue) Please follow me at BlueSky at ceesdekker.bsky.social
A major step forward toward high-resolution nanopore sequencing of full-length proteins: Molecular Cell cell.com/molecular-cell…
Single-molecule protein sequencing with nanopores! That's our latest #CDlab paper, now in Nature Reviews Bioengineering @natrevbioeng: nature.com/articles/s4422… Here Justas Ritmejeris. @XiuqiChen and myself provide an overview, with emphasis on the many recent developments.
The way towards Single-Molecule Temporal Omics, exploring the future of #nanopore in cutting-edge temporal omics @naturemethods Many thanks to Mengyin @li_mengyin and Rujun@南方科大 nature.com/articles/s4159…
.@xinshi_d about his membrane machines: set.kuleuven.be/en/news/interv…
GroEL/ES chaperonin unfolds then encapsulates a nascent protein on the ribosome biorxiv.org/content/10.110…
Harnessing DNA computing and nanopore decoding for practical applications: from informatics to microRNA-targeting diagnostics - Chemical Society Reviews pubs.rsc.org/en/content/art…
Cool concept (writing digital data into DNA epigenetically), great technical execution, and a real-world user study putting the tech into the hands of non-experts for bespoke data encoding (!?) Fun to write this News And Views. And link to original paper: nature.com/articles/s4158…
DNA is an appealing medium for data storage because of its amazing data density — a single gram can store 215,000 terabytes go.nature.com/3UoaXgR
DNA origami can create comparably giant structures, but protein design offers atomic precision, enabling molecular recognition and catalytic functions. Potential there for combining these approaches in the future. science.org/doi/10.1126/sc… @ScienceMagazine
The first paper of this very nice and productive collab with the lab of @cees_dekker Together with @XiuqiChen, Jasper van de Sande and Justas Ritmejeris took on the challenge to use nanopore reading to distinguish sulfoTyr (sY) from phosphoTyr (pY). @orc_wur @ORC_PhDs @WUR
.@baukevon, @cees_dekker & team employ single-molecule #nanopore sequencing to distinguish sulfation & phosphorylation #PostTranslationalModifications on the plant peptide hormone phytosulfokine (PSK). @KavliFoundation @tudelft #OpenAccess: go.acs.org/bja

Introducing an upgraded Claude 3.5 Sonnet, and a new model, Claude 3.5 Haiku. We’re also introducing a new capability in beta: computer use. Developers can now direct Claude to use computers the way people do—by looking at a screen, moving a cursor, clicking, and typing text.

Collaborative work of @slavicaPD, @Hashemyaser4, Jovanovic Lab and @DjuranovicLab - a new method to isolate ribosomes and translational material in minutes. We show multiple usage of our RAPPL method in biochemical and structural assays with ribosomes. Please be free to contact…
New paper alert! We used optical tweezers to resolve heterogenous folding/unfolding pathways of repeat expanded huntingtin mRNA, revealing how hairpin slippage can seed inter-strand base pairing and promote aggregation. Congrats @NothinBut_Brett! 👏👏 nature.com/articles/s4146…
Phosphorylation can regulate protein function via diverse mechanisms, including the regulation of protein conformations. In our new preprint @m_correa_m compared structures of the same proteins and protein familes in phospho and non phosphorylated forms biorxiv.org/content/10.110…
#nanopore #sequencing detects sulfation #PTM on a peptide hormone with single-molecule precision! Distinguishes sulfation from phosphorylation. By @XiuqiChen, @cees_dekker & team Read it here 🔗 go.acs.org/bhu

Easy and accurate protein structure prediction using ColabFold | Nature Protocols nature.com/articles/s4159…
United States Trends
- 1. Madison 81,8 B posts
- 2. #schoolshooting 7.874 posts
- 3. #MyMastermind 1.798 posts
- 4. Electoral College 12,6 B posts
- 5. $CUTO 10,1 B posts
- 6. For Good 509 B posts
- 7. Sayin 110 B posts
- 8. Abundant Life Christian School 37,6 B posts
- 9. Superman 164 B posts
- 10. Air Noland 1.013 posts
- 11. #GizZzVeİskaOlurMu 1.237 posts
- 12. Christmas Matters 1.078 posts
- 13. Cutoshi 2.919 posts
- 14. Constitutional Amendment 3.694 posts
- 15. Dave Clawson N/A
- 16. Ademola Lookman 107 B posts
- 17. Senate Democrats 8.244 posts
- 18. Sacramento State N/A
- 19. USPS 21,4 B posts
- 20. #BeyondTheGates N/A
Who to follow
-
 Darby Kozan
Darby Kozan
@d1kozan -
 Christina McNerney
Christina McNerney
@_chrisT3na -
 Jingliang Simon Zhang
Jingliang Simon Zhang
@simonational -
 Stephanie Yan
Stephanie Yan
@smyan__ -
 Peter DeFord
Peter DeFord
@p_deford -
 Kevin Rhine
Kevin Rhine
@KevinRhine_ -
 Merlin Li
Merlin Li
@supmerlin -
 Taylar Mouton
Taylar Mouton
@MoutonTaylar -
 Diego Rivera Gelsinger
Diego Rivera Gelsinger
@tehhoyen -
 Margaret Starostik
Margaret Starostik
@MRStarostik -
 Trina Schroer
Trina Schroer
@taschroer -
 Maryam Yamadi
Maryam Yamadi
@maryamyamadi -
 Radhika Jangi
Radhika Jangi
@radhikajangi -
 Caitlin Pozmanter
Caitlin Pozmanter
@CaiPoz
Something went wrong.
Something went wrong.