Ryan Collins
@RyanLCollins13Avid genome spelunker | Postdoc @VanAllenLab at @DanaFarber | PhD '22 @HarvardMed, @talkowskilab at @CGM_MGH, @BroadInstitute | AB @dartmouth ‘13
Similar User

@ksamocha

@TalkowskiLab

@dalygene

@jkpritch

@theFourier2k

@HeidiRehm

@ceclindgren

@AnneOtation

@genetisaur

@alexisjbattle

@gabecasis

@nickywhiffin

@mehurles

@tuuliel_lab

@hilsomartin
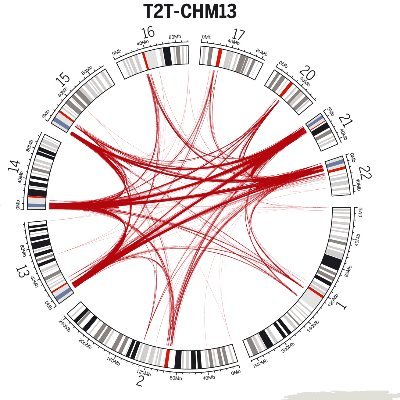
🚨🔊Interested in structural variants? Genome sequencing? Pediatric cancers? Germline variants+cancer risk? Rare variants+rare diseases? If so (or even if not!), I'm delighted to share the first paper from my postdoc @VanAllenLab @DanaFarber: biorxiv.org/content/10.110… 1/12


Excited to share this discovery: Huntington's disease is a DNA process for almost all of a cell's life. Inherited HD alleles are innocuous, just unstable – CAG repeats slowy expand throughout life, acquiring toxicity only when quite long (>150 CAGs). biorxiv.org/content/10.110…
gnomAD 4.1 is now live! This release fixes the AN issue in #gnomAD v4.0 & adds 2 new functionalities: 1) Joint AN across all called sites in exomes and genomes 2) A flag indicating when exomes and genomes frequencies are highly discordant Learn more at broad.io/gnomad_v4-1
1/ We’re excited to share our new preprint “Dissecting the contribution of common variants to risk of rare neurodevelopmental conditions” available on medRxiv! medrxiv.org/content/10.110…
We are happy to announce the release of DECIPHER dosage sensitivity tracks for human assemblies GRCh38/hg38 and GRCh37/hg19, displaying a cross-disorder dosage sensitivity map of the human genome. Learn more about this release: genome.ucsc.edu/goldenPath/new…
Yet again--to nobody's surprise--@JShendure & co blow several fields'-worth of minds in a single paper 🤯🤯 Remarkable combination of cool tech + important but understudied problem Tech like this will transform our understanding of how SVs impact cellular phenotypes & fitness!
Multiplex generation and single cell analysis of structural variants in a mammalian genome biorxiv.org/cgi/content/sh… #biorxiv_genomic
What if we could inducibly create thousands of structural variants and ecDNAs in human cells and see which ones survive? biorxiv.org/content/10.110… I’m excited to share our story on scrambling the human genome by combining prime editing, repetitive elements, and recombinases. A🧵
Oops! Samtools 1.19.1 had a bug which broke filtering on unordered BED files. This is now fixed in release 1.19.2. See htslib.org/download/ for links to tarballs and release notes.
Joint postdoc opportunity with @MarkELindsay and me based @broadinstitute! Come join us!
United States Trends
- 1. Cowboys 47,8 B posts
- 2. #WWERaw 56 B posts
- 3. Texans 59,1 B posts
- 4. Cooper Rush 8.238 posts
- 5. Pulisic 16,7 B posts
- 6. Mixon 12,2 B posts
- 7. #USMNT 3.127 posts
- 8. Aubrey 14,7 B posts
- 9. #HOUvsDAL 6.615 posts
- 10. Turpin 2.983 posts
- 11. #AskShadow 5.770 posts
- 12. Trey Lance 1.503 posts
- 13. Jake Ferguson 1.403 posts
- 14. Bruins 4.911 posts
- 15. Bray 3.852 posts
- 16. #90dayfiancetheotherway 1.813 posts
- 17. Derek Barnett N/A
- 18. Zach Lavine 2.026 posts
- 19. Weah 2.105 posts
- 20. Stingley N/A
Who to follow
-
 Kaitlin Samocha
Kaitlin Samocha
@ksamocha -
 Talkowski Lab
Talkowski Lab
@TalkowskiLab -
 Mark Daly
Mark Daly
@dalygene -
 Jonathan Pritchard
Jonathan Pritchard
@jkpritch -
 Monkol Lek
Monkol Lek
@theFourier2k -
 Heidi Rehm
Heidi Rehm
@HeidiRehm -
 Cecilia Lindgren
Cecilia Lindgren
@ceclindgren -
 Anne O'Donnell-Luria
Anne O'Donnell-Luria
@AnneOtation -
 Alicia Martin
Alicia Martin
@genetisaur -
 Alexis Battle
Alexis Battle
@alexisjbattle -
 Goncalo Abecasis
Goncalo Abecasis
@gabecasis -
 Nicky Whiffin
Nicky Whiffin
@nickywhiffin -
 Matt Hurles
Matt Hurles
@mehurles -
 Lappalainen Lab
Lappalainen Lab
@tuuliel_lab -
 Hilary Martin
Hilary Martin
@hilsomartin
Something went wrong.
Something went wrong.