Similar User

@WestermannLab

@katwandu

@JoergLab

@SharmaLab1

@CarmenAguilar_J

@Milan_Gerovac

@Chunlei_Jiao

@CMichaux13

@MoualiYoussef

@_Daniel_Ryan_

@LBarquist

@AE_Saliba

@mona_alzheimer

@jakobjung

@Jens_Hoer
2 is better than 1 – Our recent work in #mBio identifies a FoxJ as a 2nd σE-dependent #sRNA in #Fusobacterium. Not only does FoxJ act as negative regulator but also positively regulates some mRNA targets - offering a new trick for σE-dependent sRNAs. tinyurl.com/427rpkpj

Small RNA w/big roles in colonization @MoualiYoussef @StrowigLab characterize RNA landscape of Segatella copri. ID small RNA regulator essential for colonization & whose expression is regulated by metabolism of dietary components by S. copri cell.com/cell-host-micr…

Check out the spectacular work led by @MoualiYoussef - giving insights into #sRNA regulation in the unexplored #Segatella/#Prevotella impacting metabolism and interactions with the host & commensals in the gut🦠
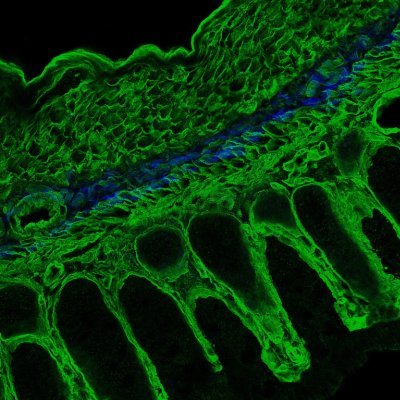
Happy to share our work, now out in @cellhostmicrobe! We investigated the human gut commensal Segatella copri (former Prevotella copri) RNA landscape and identified a small RNA essential for gut colonization. cell.com/cell-host-micr…
New research from @BullmanLab /@CJohnstonLab1, with superstar 1st author @mzepedar, out now in @Nature pinpoints “high-risk” subtype of Fusobacterium nucleatum that dominates the colorectal cancer (CRC) niche in human patient's Link : rdcu.be/dBUjH Tweet🧵below:

A new study from Bullman and @CJohnstonLab1 @fredhutch out today @Nature We identify a 'high-risk' clade of F. nucleatum that dominates the human CRC niche. Huge congrats to the amazing first author @mzepedar and the incredible team. 🧵 from Chris 👇 nature.com/articles/s4158…
New research from @BullmanLab /@CJohnstonLab1, with superstar 1st author @mzepedar, out now in @Nature pinpoints “high-risk” subtype of Fusobacterium nucleatum that dominates the colorectal cancer (CRC) niche in human patient's Link : rdcu.be/dBUjH Tweet🧵below:

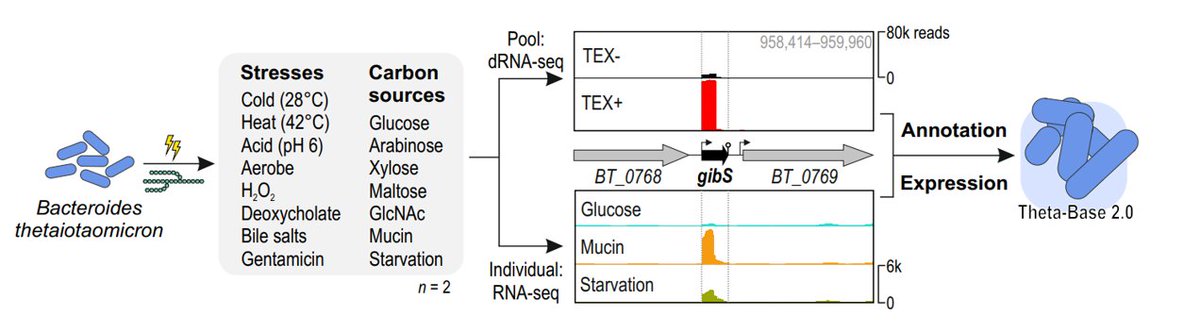
1/5: Molecular intricacies of gut bacteria! 🧬 Our study in @NatureMicrobiol , with colleagues from @Helmholtz_HIRI, @DeutschbauerLab , & @LBarquist groups, unveils B. thetaiotaomicron's transcriptional landscape across 15 gut conditions. nature.com/articles/s4156…
Very excited to finally see our study of global RNA stability in Salmonella out today in @PNASnews, where we use Bayesian statistical modeling to show bacterial transcripts have subminute half-lives, far shorter than previously thought. pnas.org/doi/10.1073/pn… 1/n
Another great collaboration with @Axel_R_Huber, @Cayetanopm, @JoskeUbels, @HansClevers, @BoxtelLab and many others. Random forest trained with #organoid #colibactin mutations => identification of pks-linked mutations in every 8th #colorectal tumor genome! authors.elsevier.com/sd/article/S15….
We (@Cayetanopm,@JensPuschhof @JoskeUbels, @HansClevers, @BoxtelLab and all others ) are happy to share our new study on colibactin-mutagenesis in organoids and distinctive features in colorectal cancer doi.org/10.1016/j.ccel…. #Cancer #Organoids #Microbiome
Happy to share our latest work, spearheaded by @prezzag in collaboration with @BeiselLab @Helmholtz_HIRI and just out @PNASNews We set up a CRISPRi system in B. thetaiotaomicron and used it to screen sRNAs for their functions. [1/5] pnas.org/doi/10.1073/pn…
#Fusobacterium harboring KH-domain containing #RBP - have a look at this great work led by @yanzhu0329 putting great effort into establishing a broadly applicable pulldown method for RBP identification using #sRNAs & uncovering an ethanolamine-dependent phenotype for KhpB/A
So excited to share that our preprint discovering KhpA/KhpB as broad RBPs in fusobacteria are now out in @NAR_Open Big thanks to my supervisor @JoergLab and @AnkeSparmann, TeamFuso @PonathF , as well as all supports from Vogel Lab @Helmholtz_HIRI! academic.oup.com/nar/advance-ar…
Proud to present SHIFTR, a novel platform for region-specific RNA interactome discovery developed by fantastic PhD student @jensaydin 👨🔬 in my group, now published in @NAR_Open: 1/8 academic.oup.com/nar/advance-ar…
A new preprint of my PhD project @JoergLab working with TeamFuso @PonathF and Valentina Cosi, identifying two KH domain proteins-KhpA and KhpB as broad RBPs in Fusobacterium nucleatum.
A global survey of small RNA interactors identifies KhpA and KhpB as major RNA-binding proteins in Fusobacterium nucleatum biorxiv.org/cgi/content/sh… #biorxiv_micrbio
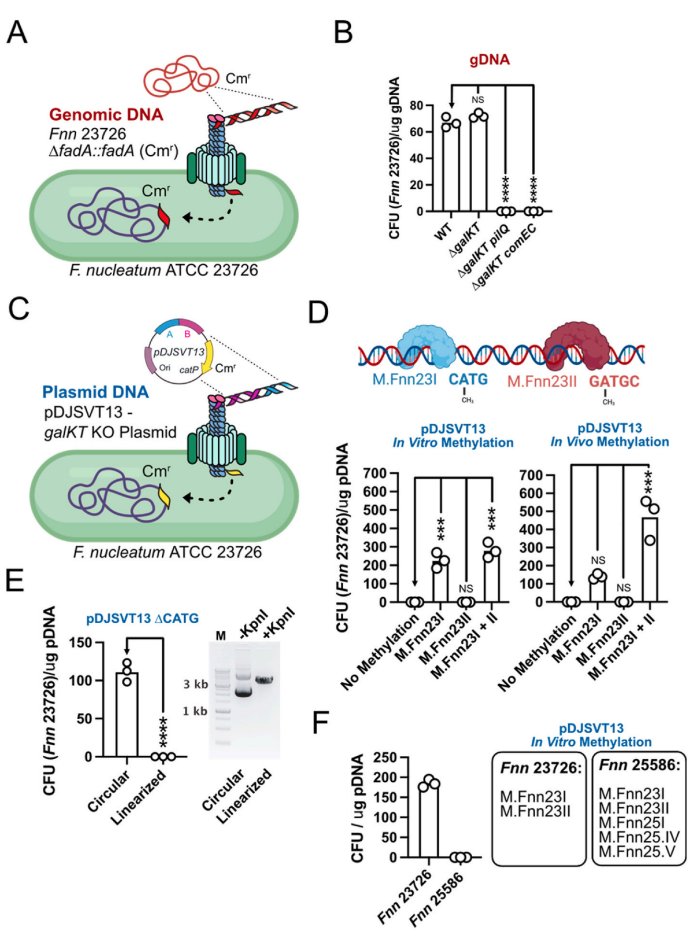
Happy to share our latest work: Fusobacterium are naturally competent through Type IV pili. Great, concise study led by @blake_e_sanders and others not on twitter. We discovered it, now you go figure out what to do with it. sciencedirect.com/science/articl…
A conveyor belt of DCs: FLT3L regulates the spatiotemporal cascade of DC-development in LNs. Fascinating work from @Kamulab @ugur_milas out @ImmunityCP, great teamwork @Wue_SI @dominic_grun @AE_Saliba - open access cell.com/immunity/fullt…
I ventured into the new world of RNA at @JoergLab with great support from @HoebartnerLab @LBarquist @jakobjung & the other co-authors! Preprint: A side-by-side comparison of peptide-delivered antisense antibiotics employing different nucleotide mimics biorxiv.org/content/10.110…
First paper from the Adams lab @NICHD_NIH is out @NatureComms! We comprehensively mapped RNA and characterized transcription termination in the Lyme pathogen B. burgdorferi — hundreds of truncated transcripts and potential regulators! nature.com/articles/s4146…
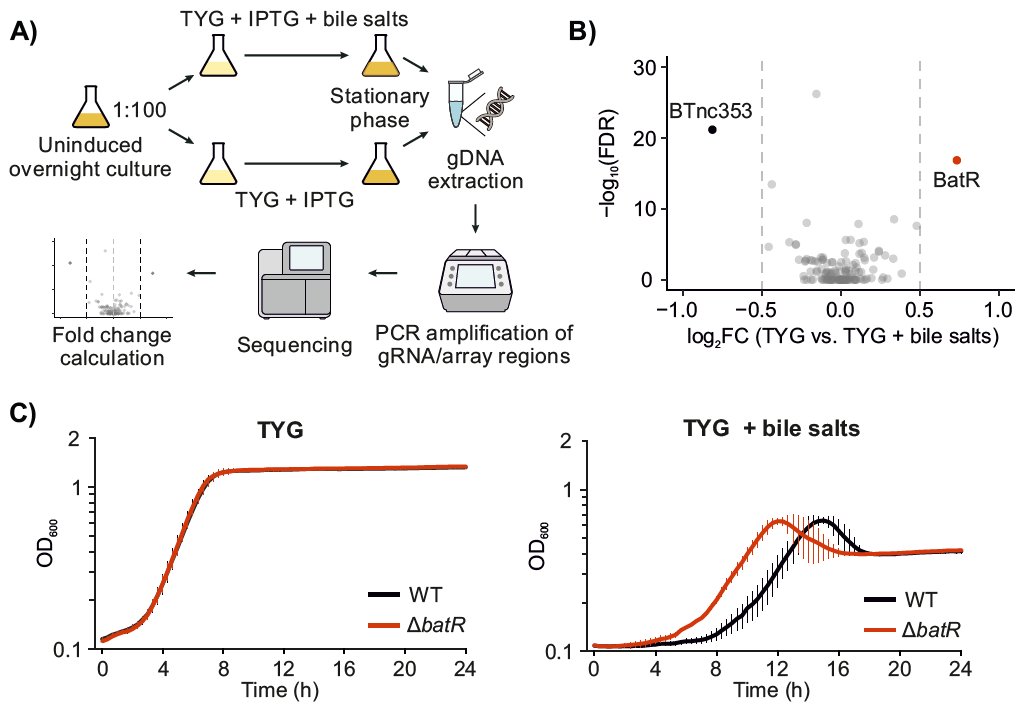
Our latest work, from @prezzag and in collaboration with @BeiselLab: we constructed a CRISPRi library of the intergenic sRNAs of B. theta and used it to identify modulators of growth under bile. Presenting BatR, the third sRNA we have characterized so far! biorxiv.org/content/10.110…
United States Trends
- 1. Kendrick 524 B posts
- 2. #AskShadow 17,4 B posts
- 3. $CUTO 7.148 posts
- 4. Luther 39,3 B posts
- 5. Drake 75,2 B posts
- 6. Daniel Jones 45 B posts
- 7. Wayne 51,9 B posts
- 8. MSNBC 179 B posts
- 9. Kdot 5.990 posts
- 10. Squabble Up 23,5 B posts
- 11. TV Off 32,2 B posts
- 12. Giants 77,3 B posts
- 13. Dodger Blue 11,6 B posts
- 14. Reincarnated 30,5 B posts
- 15. Kenny 24 B posts
- 16. #BO6Sweepstakes N/A
- 17. NASA 69,3 B posts
- 18. Gloria 45,9 B posts
- 19. #TSTTPDSnowGlobe 4.961 posts
- 20. One Mic 4.699 posts
Who to follow
-
 Westermann lab
Westermann lab
@WestermannLab -
 Katharina Wandera
Katharina Wandera
@katwandu -
 Jörg Vogel
Jörg Vogel
@JoergLab -
 Sharma Lab
Sharma Lab
@SharmaLab1 -
 Carmen_Aguilar
Carmen_Aguilar
@CarmenAguilar_J -
 Milan Gerovac
Milan Gerovac
@Milan_Gerovac -
 Chunlei Jiao
Chunlei Jiao
@Chunlei_Jiao -
 Charlotte Michaux
Charlotte Michaux
@CMichaux13 -
 Youssef El Mouali
Youssef El Mouali
@MoualiYoussef -
 Daniel
Daniel
@_Daniel_Ryan_ -
 Lars Barquist
Lars Barquist
@LBarquist -
 Emmanuel Saliba
Emmanuel Saliba
@AE_Saliba -
 Mona Alzheimer
Mona Alzheimer
@mona_alzheimer -
 Jakob Jung
Jakob Jung
@jakobjung -
 Jens Hör
Jens Hör
@Jens_Hoer
Something went wrong.
Something went wrong.