Lutimba Stuart
@LutimbaStuartGraduate Teaching Assistant | London South Bank University Bioinformatics and Statistics Structural Computational Biology
Similar User

@guglielmo_ted

@Tatiana_ATPase

@gchrisjenkins

@asmolsim

@HOUDAIFAMUSLIM

@gouravchoudhir

@PallavSengupta3

@Kayenat__

@SuryaSingh85
Am so happy that our paper just got accepted and published in Nature Communications @NatureComms 🤩🤩🤩🤩🤩🤩🤩🤩🤩🤩🤩🤩 nature.com/articles/s4146…
With David and the Baker Lab in the spotlight today, I wanted to share some insights into the @UWproteindesign and how it operates, a glimpse behind the curtain. I had planned to write this post-graduation, but now seems as good a time as any. (Got twitter blue free trial so this…
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”

AlphaFold structures might be as good as experimental structures for virtual screening

AlphaFold2 structures template ligand discovery biorxiv.org/cgi/content/sh… #bioRxiv
Interested to contribute to research addressing #Theory and Methods for Non-equilibrium Atomistic #Simulations of Complex #Biomolecules? Join the #grubmüller lab and apply for #PhD or #Postdoc positions @mpi_nat in Göttingen 🇩🇪 #Jobs #AcademicChatter 🔗 mpinat.mpg.de/4588467/60-23?…
Do you know you can contribute to the GROMACS code? Just need to enter #GROMACS #gitlab, and check the #issues! Choose the one you like and start! Have questions? Attend gmx bi-weekly meetings! :) #gmx #GROMACS #developer #compchem

“Guidelines for Reporting Molecular Dynamics Simulations in JCIM Publications” Interesting (and somewhat controversial) because it both gives guidelines for reporting, but also on how simulations should be setup, performed, and analysed. doi.org/10.1021/acs.jc…
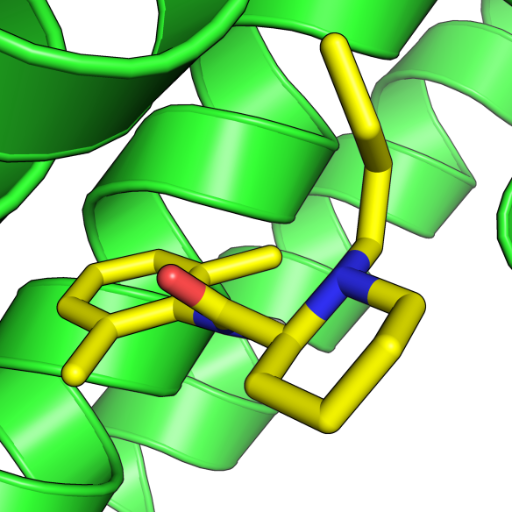
Me: Can I have a #structure of my #protein of interest? Alphafold: We have it in the #database, no need to make new one. Structure in the database:

We are organizing a free symposium on "Computational methods in drug and materials design" with @eriklindahl @BioExcelCoE and Tom Cheatham as our plenary speakers. In-person and online details below. #compchem
RT! We are organizing a Symposium on “Computational methods in drug and materials design” at @BRFAA_IIBEAA Amphitheater, on Monday 20th June, 3-7pm CET. Check the program here: allodd-itn.eu/uploads/9/4/6/… For those not in Athens connect via zoom: zoom.us/j/98001118799?… #compchem


Glad to share that we are organizing our second workshop on DeepModeling for molecular simulation. Free and fully virtual. Dates: July 7-8. More info ccsc.princeton.edu/upcoming-event… and registration link forms.gle/vULjA7abwgGZUw…. Please share!

Coursera had started with my machine learning class. I’m delighted to announce that I’ll soon be teaching a new Machine Learning Specialization, created by @StanfordOnline & @DeepLearningAI_ Please help me spread the word! You can sign up here: deeplearning.ai/program/machin…
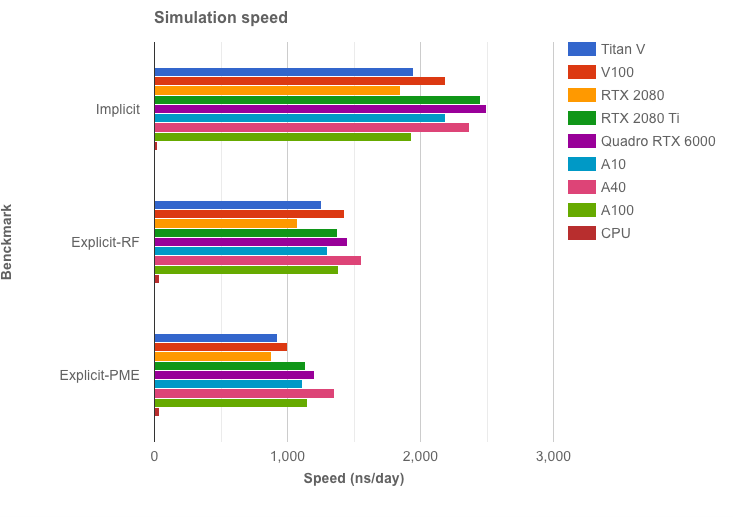
OpenMM 7.7 benchmarks are up! 🔥🔥🔥 DHFR with PME gets 1.35 microseconds/day on an @nvidia A40! Check out the benchmarks yourself here: openmm.org/benchmarks
Registration is open for the @cecamEvents workshop “20 years of Metadynamics”. September 5-8 2022, Lausanne (CH). Organized by @GroupParrinello, Alessandro Laio, @BussiGio, @gervasiolab, @evi_gkeka, and myself #compchem #cecam #compBio More info here: cecam.org/workshop-detai…
Omicron variant’s spike protein of SARS-CoV-2, as well as of the original virus’ spike protein at a resolution of 2Å - the highest resolution obtained as yet. Congrats Didier Trono biorxiv.org/content/10.110…
MoleGuLAR: Molecule Generation Using #ReinforcementLearning with Alternating Rewards #compchem pubs.acs.org/doi/10.1021/ac… @manan_goel @siddharthalsd @deva_priyakumar #current_issue #MachineLearning #DeepLearning

Dependence between force and computed unbinding rates. It is interesting that the streptavidin-biotin system does not clearly follow Bell’s law, but this system is probably an exception because of the high affinity. #compchem arxiv.org/abs/2112.03249
Here is another excellent starting point: @AndreasBenderUK sciencedirect.com/science/articl…

RT! We are organizing a Symposium on “Computational methods in drug and materials design” at @BRFAA_IIBEAA Amphitheater, on Monday 20th June, 3-7pm CET. Check the program here: allodd-itn.eu/uploads/9/4/6/… For those not in Athens connect via zoom: zoom.us/j/98001118799?… #compchem


Unveiled! Mode of enantioinduction & the key catalytic pocket (and what's in it) of heteroarene hydrogenation. It took us 5 years and scientists from several research institutes to answer this question (tricksy hobbit). Just published in @Chemical_Science! kurzelinks.de/zi5x

The art of solving problems with Monte Carlo simulations | Gabriel Carvalho bit.ly/3jzLJL2 #AI #DeepLearning #MachineLearning #DataScience

United States Trends
- 1. Kendrick 711 B posts
- 2. #AskShadow 25,7 B posts
- 3. MSNBC 235 B posts
- 4. $LEAFS N/A
- 5. Scott Bessent 6.459 posts
- 6. Drake 92,6 B posts
- 7. Luther 52,3 B posts
- 8. The Warm Up 14,8 B posts
- 9. Brandon Allen 2.348 posts
- 10. Wayne 62,8 B posts
- 11. LinkedIn 44 B posts
- 12. Kdot 10,9 B posts
- 13. TV Off 44,5 B posts
- 14. Daniel Jones 49 B posts
- 15. Dobbs 2.052 posts
- 16. NASA 74,2 B posts
- 17. Purdy 7.712 posts
- 18. Friday Night Lights 1.064 posts
- 19. Snoop 22,2 B posts
- 20. Maddow 37,4 B posts
Something went wrong.
Something went wrong.