James Holder
@JRHCellCycleHusband, father and post-doc at Uni of Oxford interested in the impact and regulation of PTMs throughout the cell cycle.
Similar User

@microtubule_guy

@vmdraviam

@AdeSaurin

@isabella__k

@CarlosConde_i3S

@JPLab_

@Draviam_LAB

@Stimac_V

@EmileRoberts97

@jstumpffVT

@KrunoVuk

@AlexT_biology

@MikecPetra

@ernatintor

@LabWestermann
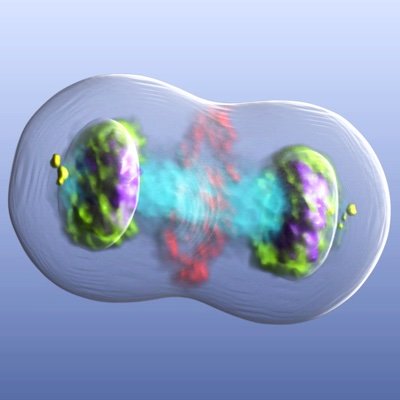
Very proud to share the new paper from myself (Gergely lab) and Jennifer Miles (@baylisslab) #cellcycle #mitosis #centrosome #kinase Lots of exciting discoveries within! embopress.org/doi/full/10.10…
A study, led by @baylisslab and @BiochemOxford has revealed new knowledge about an enzyme involved in cancer treatment. The study reveals the process of how Aurora-A binds to other proteins to perform its role in cell division. biologicalsciences.leeds.ac.uk/biological-sci…

Thank you very much to our editor @HVodermaier for expertly guiding us through the peer review process! And to the reviewers for their comments which really helped us reach the finished product.
The structure of the human Aurora-A:CEP192 complex reveals its importance in facilitating Aurora-A autophosphorylation, TPX2 binding, and activity on the mitotic spindle @JRHCellCycle, Jennifer Miles, Richard Bayliss, @fanni_gergely et al embopress.org/doi/full/10.10…

Still time to register for Wednesday’s virtual conference. Excited to share some of my work with the community!
6 breakout talks added!! Virtual Cell Cycle Club meeting: Wednesday March 13 3 Selected talks: Gergely, Mittnacht & @KabecheLab 6 talks in the Breakout Session 1 Keynote:@JanSkotheim Please REGISTER as soon as possible for this FREE event @abcam events.abcam.com/event/bfdb7a23…… 📷


I'll be presenting an exciting story focussed on spatial control of the mitotic kinase Aurora-A at the next virtual Cell Cycle conference (13th March). Very grateful to the organizers for selecting my abstract for a short talk! There is still time to register to attend.
PROGRAMME Virtual Cell Cycle Club meeting: Wednesday March 13 3 short talks: Gergely, Mittnacht & @KabecheLab Breakout session talks Keynote: @JanSkotheim Breakout slots still available, deadline this Friday!! To REGISTER or SUMIT abstract @abcam events.abcam.com/event/bfdb7a23…

Come by my poster, B174, at #cellbio2023 to learn about our exciting story surrounding Aurora A kinase. We’re very proud of the work from myself and the fantastic @SPIDR_IDP team!
Heading to Boston this weekend for #CellBio2023 . Can’t wait to share my work and meeting lots of wonderful cell biologists! Come find me at poster 1900, 13.45 on Monday to learn about my work on Aurora A at centrosomes.
Very pleased to see our @SPIDR_IDP paper on constrained N-Myc peptides out @ChemBioChem from @dawber_robert with @MWrightLab @AJWilsonGroup and @baylisslab …mistry-europe.onlinelibrary.wiley.com/doi/10.1002/cb…
Happy to have played a part in this great work spearheaded first by @SarahCarden92 and then by @ElisaVitiello . Based on a cool discovery by @dr_takashi_ochi . This really opens the doors to being able to answer some vital questions in centrosome biology.
Interested in studying centrosome composition? Check out CAPture, our 1-step purification method to obtain high-resolution centrosome proteomes from mammalian cells. cell.com/developmental-…
Very interesting new paper from one of the @SPIDR_IDP team. Great work Takashi.
My first preprint is out! This is about the identification of plant orthologs of an NHEJ protein PAXX. The first half of this paper is an evolutional analysis of NHEJ proteins in eukaryotes, and the second half is in-vitro studies of Arabidopsis PAXX biorxiv.org/content/10.110…
Thanks to everyone who came to my poster today, had some fantastic discussions! All the talks have been fantastic so far as well!
The conference “Mitotic spindle: From living and synthetic systems to theory” is open! phy.pmf.unizg.hr/~mitosis/ Follow us #SpindleCroatia2023!
Im looking to analyze some mass spec data in R, a sequence alignment of identified phosphorylations to see which are evolutionarily conserved. I am a complete beginner with R and coding. Does anyone have recommendations for where to start with biology specific tutorials for R?
The finished proofs of our paper from the Barr lab regarding control of mitotic exit are online. Now that it is published that PhD chapter of my life is officially over. Many thanks to @eLife for the well written and informative digest of the work.
Changes in protein levels during cell division reveal how the process is carefully controlled elifesciences.org/digests/59885/…
United States Trends
- 1. Mike 1,84 Mn posts
- 2. Serrano 239 B posts
- 3. Canelo 16,5 B posts
- 4. #NetflixFight 72,1 B posts
- 5. Father Time 10,7 B posts
- 6. #netflixcrash 15,8 B posts
- 7. Logan 78,7 B posts
- 8. He's 58 25,9 B posts
- 9. Rosie Perez 14,9 B posts
- 10. ROBBED 102 B posts
- 11. #buffering 10,9 B posts
- 12. Boxing 299 B posts
- 13. Shaq 16,2 B posts
- 14. My Netflix 83,1 B posts
- 15. Roy Jones 7.170 posts
- 16. Tori Kelly 5.246 posts
- 17. Ramos 69,8 B posts
- 18. Barrios 50,6 B posts
- 19. Muhammad Ali 18,4 B posts
- 20. Cedric 22 B posts
Who to follow
-
 Vladimir Volkov 🇺🇦
Vladimir Volkov 🇺🇦
@microtubule_guy -
 Viji Draviam
Viji Draviam
@vmdraviam -
 Adrian Saurin
Adrian Saurin
@AdeSaurin -
 Isabella Koprivec
Isabella Koprivec
@isabella__k -
 Carlos Conde
Carlos Conde
@CarlosConde_i3S -
 JP Lab
JP Lab
@JPLab_ -
 Viji Draviam
Viji Draviam
@Draviam_LAB -
 Valentina Stimac
Valentina Stimac
@Stimac_V -
 Emile Roberts
Emile Roberts
@EmileRoberts97 -
 Jason Stumpff
Jason Stumpff
@jstumpffVT -
 Kruno Vukušić
Kruno Vukušić
@KrunoVuk -
 Alexandre Thomas
Alexandre Thomas
@AlexT_biology -
 Petra Mikec
Petra Mikec
@MikecPetra -
 Erna
Erna
@ernatintor -
 WestermannLab
WestermannLab
@LabWestermann
Something went wrong.
Something went wrong.