Similar User

@SpringerNathan

@SchnableLab

@addie_maize

@MaizeZynskiHI

@JinliangYang

@CoopMaize

@SigmaFacto

@UMNplantbreed

@maliagehan

@MaizeGDB

@JoeGage10

@bob_vanburen

@DamonLisch

@schnablep

@stup0004
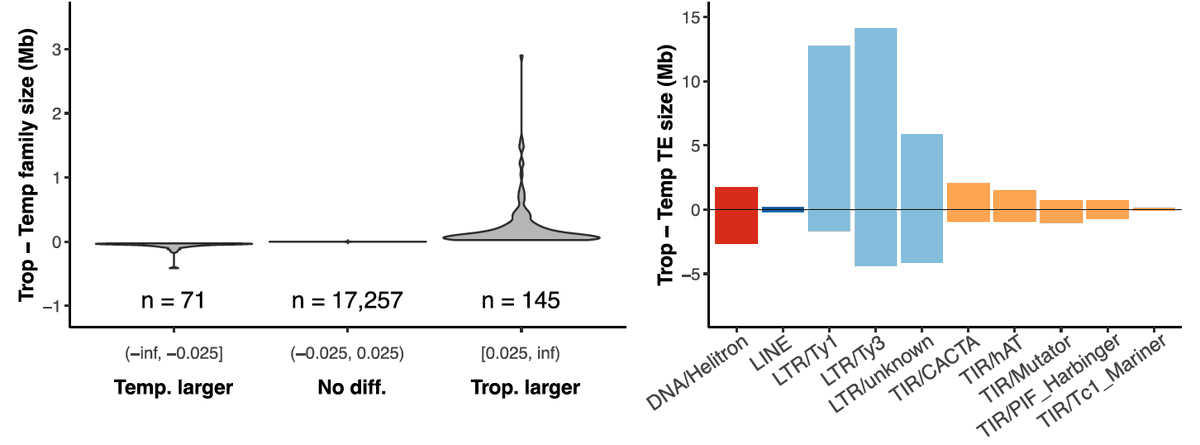
How TEs gain and lose between tropical and temperate maize inbreds? We found that tropical corns have 35Mb more TEs than temperates, and it's mainly due to a small number of LTR families that are young, less methylated, and more expressed in tropical maize.pubmed.ncbi.nlm.nih.gov/39251347/
Happy to share four strategies to annotate TEs and other repeats in maize and other plant genomes. You can shop for an approach depending on the goal, including gene-focused annotation, TE-focused study, comprehensive annotation, and pan-genome annotation: cshprotocols.cshlp.org/content/early/…
Happy to share a short review of how TEs and other repeats can contribute to biological functions in maize: cshprotocols.cshlp.org/content/early/…. Thanks to the reviewer and editorial office for all the great suggestions!
Here, Woodhouse et al (@USDA_ARS, @IowaStateU, @Mizzou) provide an overview of the key resources available at @MaizeGDB, including #maize genomes, comparative genomics, and pan-genomics tools. cshprotocols.cshlp.org/content/early/…

Congrats to Rita Hayford on the publication of her work mapping RNA-Seq reads from 24 publicly available datasets (17 abiotic & 7 biotic studies) to B73v5 and annotating over 3k stress-related functional annotation of maize gene models. doi.org/10.1186/s12864…
I am hiring a postdoc to work on evolutionary genomics projects with maize and its wild relatives. This is part of a broad collaborative project known as CERCA that aims to improve maize with adaptations from the wild. Please share or consider applying! isu.wd1.myworkdayjobs.com/IowaStateJobs/…
Excited to share AnnoSINE_v2 that can annotate SINEs in large plant and animal genomes! Available via bioconda: anaconda.org/bioconda/annos…
Accelerating de novo SINE annotation in plant and animal genomes biorxiv.org/cgi/content/sh… #biorxiv_bioinfo
Cory Hirsch, University of Minnesota "Rover Based Field Detection and Quantification of FHB" #nfhbforum23

If you are at the #ACSMtg, stop by poster 1186 on Tuesday to talk about recent developments in predicting corn chip quality traits based on NIR spectroscopy!

A new pathogen called #CornTarSpot is causing significant yield loss in MN. @josesolorzano73 et al. have published a diagnostics guide to help recognize the symptoms and signs of disease. mitppc.umn.edu/news/corn-tar-… @UMNPlantPath @UMNExt Research funded by @mnenrtf

Congratulations to @burnsmj7 on being awarded the Hayes Phillips Award! Im so proud of you and all that you have accomplished.



Congratulations to this year's Hayes Philips and Alumni Award recipients! All are welcome to join for the coffee reception and award ceremony from 10:00-10:30 AM &10:30-12:00PM, May 5th. Coffee reception will take place in the Cargill Atrium followed by the ceremony in Room 105.

.@ManishaMuna with a wonderful talk on TE polymorphisms in maize, alaso sharing her shiny app for visualing maize assembly comparisons mmunasin.shinyapps.io/nam_sv/
On my way to #MGM2023, excited to meet and talk with folks -- I'll be talking Saturday at 7:20 PM about our work looking at how TEs contribute to structural variation in inbred maize lines 🌽🌽

If you are at #MGM2023, stop by poster 198 to learn about my research developing an image analysis pipeline to quantify pericarp retention in nixtamalized maize kernels!

Attention plant science students and postdocs! Take charge of your career path and enhance your skills with our exclusive Career Development Workshop, sponsored by the Maize Genetics Cooperation (MGC) Membership Committee. iastate.qualtrics.com/jfe/form/SV_5B…

🚨 We've got a new preprint out 🚨 - We developed a series of logical frameworks for analyzing how transposable element content contributes to structural variation (SV) within maize 🌽🧬🌽. How'd we do this + what did we find?? - 1/11
Combined analysis of transposable elements and structural variation in maize genomes reveals genome contraction outpaces expansion biorxiv.org/cgi/content/sh… #bioRxiv
Plant Postdocs, are you coming to #MGM2023? 🌽🌽🌽 Please come and join the postdoc meet up on March 18th (10:00-10:40am) at the refreshment area. Use #MGMPostdocMeetUp to connect with other postdocs and we can re-tweet about your talk, poster & networking sessions.

Plant breeders use genetic information to predict how well their crops will perform. For some traits, we can improve prediction accuracy when using all available information in the plant genome, including structural variants! More on our #biorxiv preprint! biorxiv.org/content/10.110…
Our preprint describing a new model for the origin of #corn just dropped today. Preprint is here: biorxiv.org/content/10.110… And a (sort of) brief thread can be found here: ecoevo.social/@jrossibarra/1…
United States Trends
- 1. Brian Kelly 7.720 posts
- 2. Mizzou 5.873 posts
- 3. Feds 35,9 B posts
- 4. #UFC309 41,5 B posts
- 5. Louisville 6.233 posts
- 6. Gators 10,4 B posts
- 7. Nebraska 11 B posts
- 8. Luther Burden 1.128 posts
- 9. Stanford 8.870 posts
- 10. Nuss 3.265 posts
- 11. Arian Smith N/A
- 12. #MissUniverse 36,3 B posts
- 13. Brohm N/A
- 14. Napier 5.407 posts
- 15. #AEWCollision 4.961 posts
- 16. #MostRequestedLive 10,6 B posts
- 17. Lagway 4.919 posts
- 18. #Huskers 1.573 posts
- 19. Baylor 3.904 posts
- 20. Florida 71 B posts
Who to follow
-
 Nathan Springer
Nathan Springer
@SpringerNathan -
 Schnable Lab
Schnable Lab
@SchnableLab -
 Addie Thompson
Addie Thompson
@addie_maize -
 MaizeZynski Hawaii
MaizeZynski Hawaii
@MaizeZynskiHI -
Jinliang Yang
@JinliangYang -
 MaizeGeneticsCoop
MaizeGeneticsCoop
@CoopMaize -
 Shujun Ou
Shujun Ou
@SigmaFacto -
 UMN Plant Breeding Center
UMN Plant Breeding Center
@UMNplantbreed -
 Malia Gehan
Malia Gehan
@maliagehan -
 MaizeGDB
MaizeGDB
@MaizeGDB -
 Joe Gage
Joe Gage
@JoeGage10 -
 Robert VanBuren
Robert VanBuren
@bob_vanburen -
 Damon Lisch
Damon Lisch
@DamonLisch -
 Pat Schnable
Pat Schnable
@schnablep -
 Bob Stupar
Bob Stupar
@stup0004
Something went wrong.
Something went wrong.