Haedong Kim
@HaedongKPostdoc with @JShendure @UWgenome. WRF postdoctoral fellow. Interested in developing and applying high-throughput approaches to study gene regulations.
Similar User

@connorkub

@XiaoyiLi10

@_Choi_Junhong

@JennyNathans

@VinetteY

@wchenomics

@junyue_cao

@FloChardon

@sudpinglay

@CXchengxiangQIU

@Nobu_Hamazaki

@sdomcke

@aishwaryagogate

@LaraMuffley

@TroyMcDiarmid
Check out our recent back-to-back papers on how human DICER produces microRNAs in cells! (Really thank @MeisterLab for this beautiful N&V!)
MicroRNA uses a gym to get fit for cuts by Dicer enzyme. A terrific N&V by @MeisterLab discusses new discoveries about how Dicer functions. go.nature.com/3lT1EHj
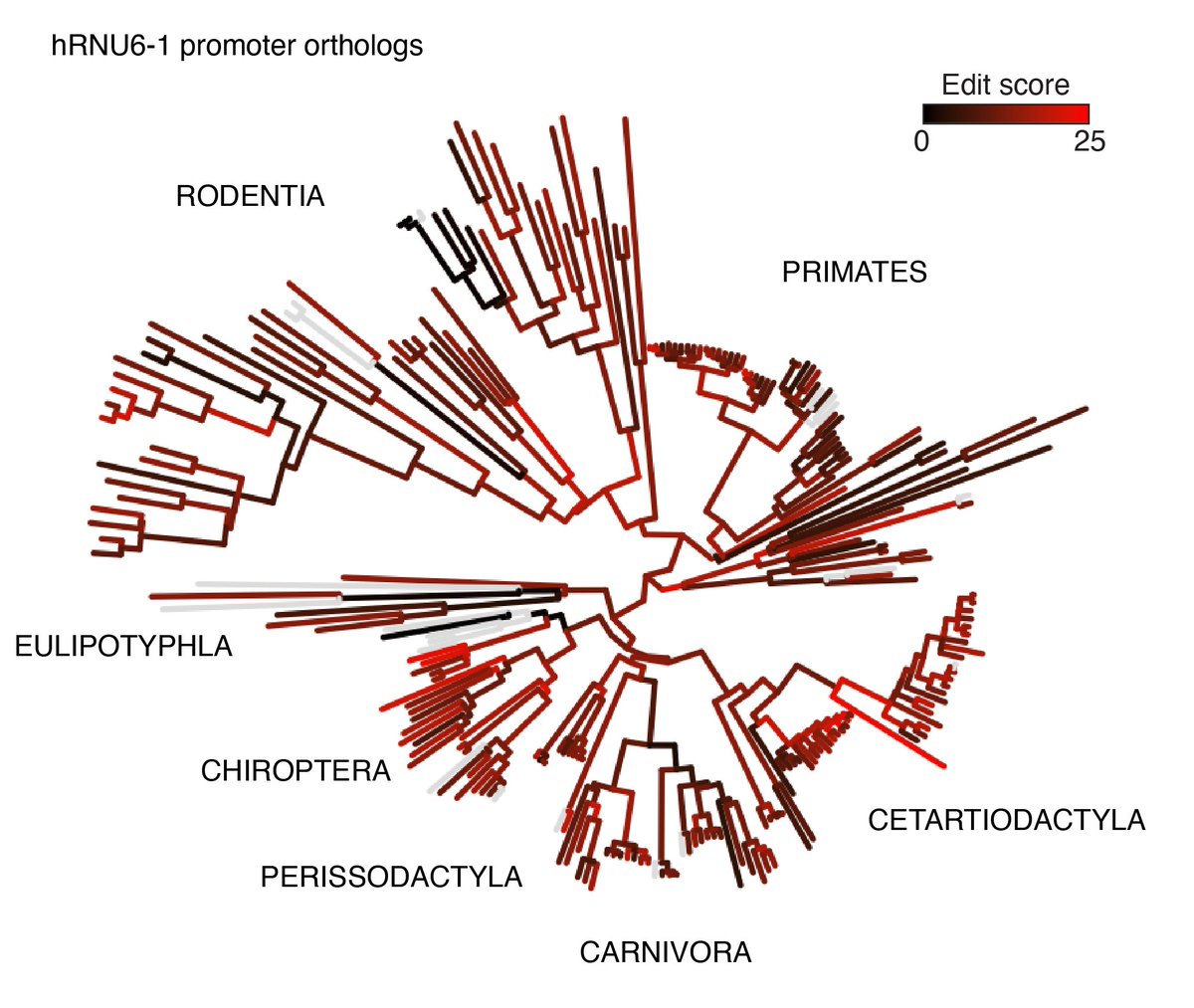
Genome engineering excelled at small nucleotide changes, but large edits remained challenging. Recent innovations are changing this. We reviewed the field of structural variant engineering @NatureGenet 🧬 @JulianeWeller @TVanderstichele @LeopoldParts nature.com/articles/s4158…
Excited to share my PhD work in @Agnelsfeir’s lab describing a method to engineer mitochondrial DNA deletions in human cells and our exploration of how cellular metabolism and transcription respond to deletion heteroplasmy. biorxiv.org/content/10.110… 1/19
Beyond stoked to share our latest, entitled “Diversified, miniaturized and ancestral parts for mammalian genome engineering and molecular recording” ! biorxiv.org/content/10.110…
Very happy to share our latest w/ @dschweppe1 and @Nobu_Hamazaki labs, where we map the temporal dynamics and proteomic landscape of mammalian gastruloids. Sneek Peak 🧵below #stembryo #gastruloid #proteomics biorxiv.org/content/10.110…
Our paper ‘Whole-embryo Spatial Transcriptomics at Subcellular Resolution from Gastrulation to Organogenesis’ is now on biorxiv! We introduce weMERFISH, a platform for profiling gene expression in entire embryos at subcellular resolution. Here is🧵biorxiv.org/content/10.110… (1/12)
Excited to share my postdoc work from the @schierlab This project was a joint effort with many collaborators @seeliglab, @castillohair,Lucia Du, @YiqunWang3,@adamcarte,@mcolomerr, Chris Yin. 1/20 biorxiv.org/content/10.110…
Truly amazing work!! Congrats to all involved!!
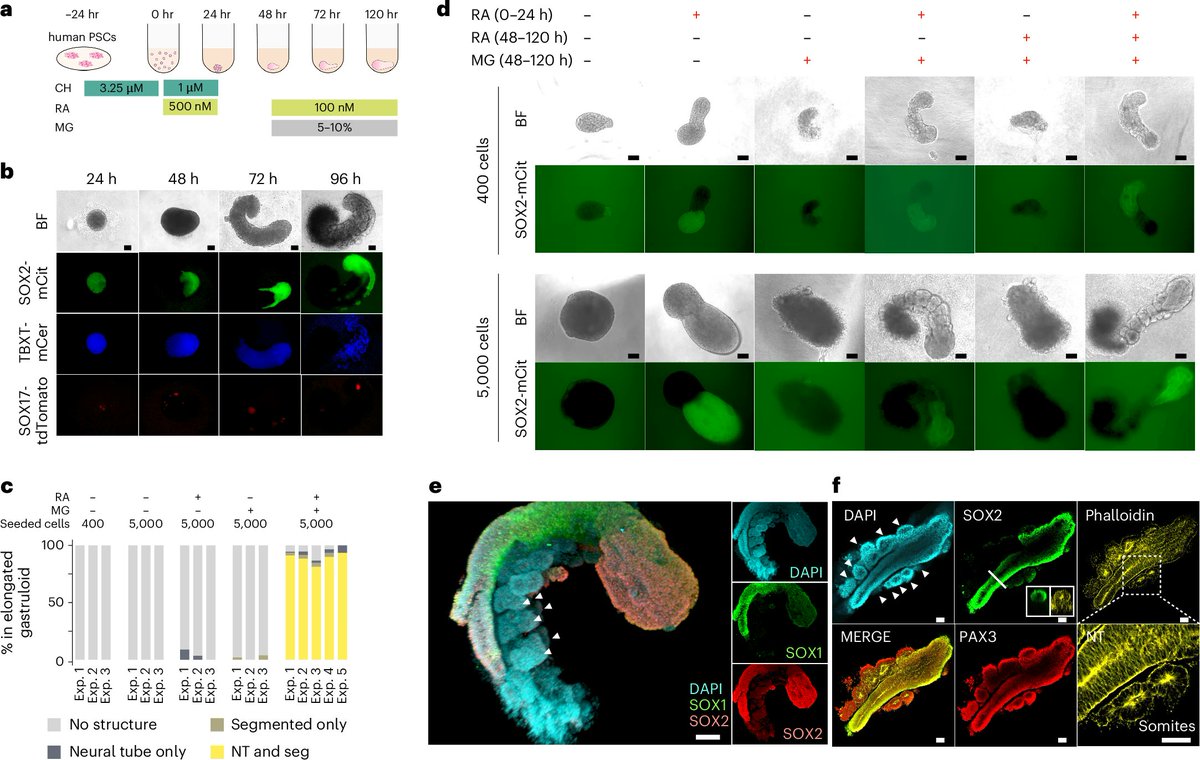
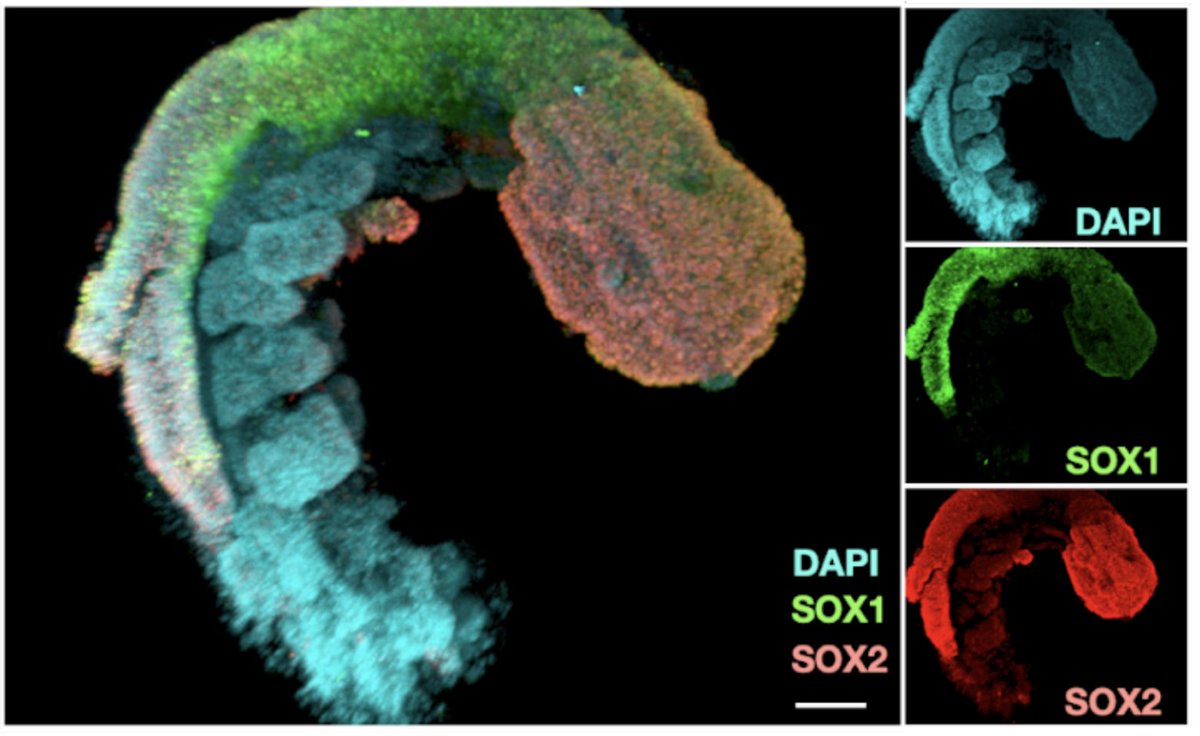
Excited to share our human RA-gastruloid model in @NatureCellBio! Early retinoic acid supplementation, followed by later Matrigel addition, robustly induces human embryonic morphologies and diverse cell lineages, including somitic, renal, cardiac, and neural cell types in…
Today out in @biorxivpreprint we present SCOPE. An optics-free, self-registering array of DNA encoded beads for spatial genomics. This was work led by the intrepid duo of Hanna Liao and Sanjay Kottapalli, with @JShendure's lab. biorxiv.org/content/10.110…
Super excited to share that ENGRAM is out today! ENGRAM cells are programmed to write their histories into the genome, recording the intensity, duration, and order of biological events simultaneously. nature.com/articles/s4158…
Our paper exploring the effect of chromatin on prime editing and diverse pegRNA designs was published today! 👨💻🧬🎉 rdcu.be/dLu0f pridict.it @UZH_Science @NatureBiotech @schwanklab @krauthammerlab @bvansteensellab 🧵

Thrilled to share the culmination of 4 years of #Fiberseq research enabling the accurate quantification of chromatin accessibility across the 6 Gbp diploid human genome with single-molecule and single-nucleotide precision. See thread for the main findings biorxiv.org/content/10.110…
Our DNA Typewriter protocol (written by @LiaoHanna, @JShendure, and myself) is out! Thank you @NatureProtocols for writing/editing this with us and having it as this week’s #FeaturedProtocol :-)
#FeaturedProtocol this week from Jay Shendure on using #DNATypewriter (#primeediting-based method) for tracing insertional events bit.ly/4cegahU
Please take a look at this amazing research about single-cell reporter assays!
After a constructive set of reviews, our single-cell reporter work is OUT nature.com/articles/s4159… Check out the briefing if you want the tldr nature.com/articles/s4159… I am particularly fond of some of the new applications. Mini-update.
Congrats, Jenny!
Excited to share a review of lineage & molecular recording methods w/ potential applications in cutaneous biology in @theJIDJournal Thank you Jessica Ayers @CorySimpsonMD @JShendure for their support conceptualizing & writing! jidonline.org/article/S0022-…

Amazing work led by an amazing scientist! Congrats, Xiaoyi!!
Our work on studying chromatin context-dependent regulation of prime editing is officially out on @Cell cell.com/cell/fulltext/…
Very cools papers about how to enable gene dosage-invariant expressions using a miRNA-based circuit!
Delivering and expressing a gene in cells is usually a messy (heterogeneous) process. The messiness interferes with research and applications such as gene therapy. In two new papers, we introduce a toolbox of synthetic miRNA-based control circuits that enable more precise, gene…

Our paper on lineage motifs is now online at @Dev_Cell 😀 We hope that this approach will be widely useful for anyone interested in analyzing tree-type datasets!
Online now: Lineage motifs as developmental modules for control of cell type proportions dlvr.it/T2lKM6
Congrats to all involved! Really amazing work!
Our latest out today in @Nature We profiled 12 million single cells from mouse embryos spanning gastrulation to birth, defined cell type tree from zygote to birth, and unexpectedly found crazy fast changes within first hour of extrauterine life. OA PDF: rdcu.be/dyDAg
We (with @VinetteY and @JShendure) are thrilled to announce our new paper on the "human RA-gastruloid" model, showcasing morphologies and transcriptomes akin to E9.5 (mouse) or CS10-11 embryos (biorxiv.org/content/10.110…). (1/n)
Structural variants (SV) impact more nucleotides per human genome than other forms of genetic variation. To better understand SV impact, we developed a method to 'shuffle' mammalian genomes and characterize them with single-cell resolution in a pool. biorxiv.org/content/10.110…
United States Trends
- 1. Dodgers 49,5 B posts
- 2. Lakers 37,9 B posts
- 3. #DWTS 74,4 B posts
- 4. Chandler 39,4 B posts
- 5. Dylan Harper 1.556 posts
- 6. Duke 38,1 B posts
- 7. #kaicenat 4.632 posts
- 8. Ilona 12,9 B posts
- 9. Joey 27,2 B posts
- 10. Sasaki 8.878 posts
- 11. Ohtani 6.988 posts
- 12. Cooper Flagg 5.044 posts
- 13. Matt Allocco N/A
- 14. Soto 25,7 B posts
- 15. Beal 7.172 posts
- 16. #IslamabadMassacre 391 B posts
- 17. Glasnow 2.926 posts
- 18. Suns 18,8 B posts
- 19. Kershaw 1.693 posts
- 20. Buehler 1.305 posts
Who to follow
-
 Connor Kubo
Connor Kubo
@connorkub -
 Xiaoyi Li
Xiaoyi Li
@XiaoyiLi10 -
 Junhong Choi
Junhong Choi
@_Choi_Junhong -
 Jenny Nathans
Jenny Nathans
@JennyNathans -
 wei yang
wei yang
@VinetteY -
 Will Chen
Will Chen
@wchenomics -
 Junyue
Junyue
@junyue_cao -
 Flo Chardon
Flo Chardon
@FloChardon -
 Sudarshan Pinglay
Sudarshan Pinglay
@sudpinglay -
 Chengxiang Qiu
Chengxiang Qiu
@CXchengxiangQIU -
 Nobu Hamazaki
Nobu Hamazaki
@Nobu_Hamazaki -
 Silvia Domcke
Silvia Domcke
@sdomcke -
 Aishwarya Gogate
Aishwarya Gogate
@aishwaryagogate -
 Lara Muffley
Lara Muffley
@LaraMuffley -
 Troy McDiarmid
Troy McDiarmid
@TroyMcDiarmid
Something went wrong.
Something went wrong.