Stefan Engelen
@stefan_engelenBioinformatician at CEA/Genoscope France
Similar User

@fr_genomics

@BioinfoGenotoul

@vallenet

@J_M_Aury

@GeT_Genotoul

@INRAE_Genomics

@GenoLAGE

@GenoLabgem

@GUILLAUMEGAUTRE

@EPGV_INRAE

@pholive81

@kevinlebrigand

@ch_baum

@olivierlucas29

@agroppi
BoardION: real-time monitoring of Oxford Nanopore sequencing instruments ift.tt/3eMJyAR #bioinformatics
Happy to share BoardION, a web application for real time monitoring of sequencing experiments from @nanopore devices, @BMC_Bioinforma ift.tt/3eMJyAR

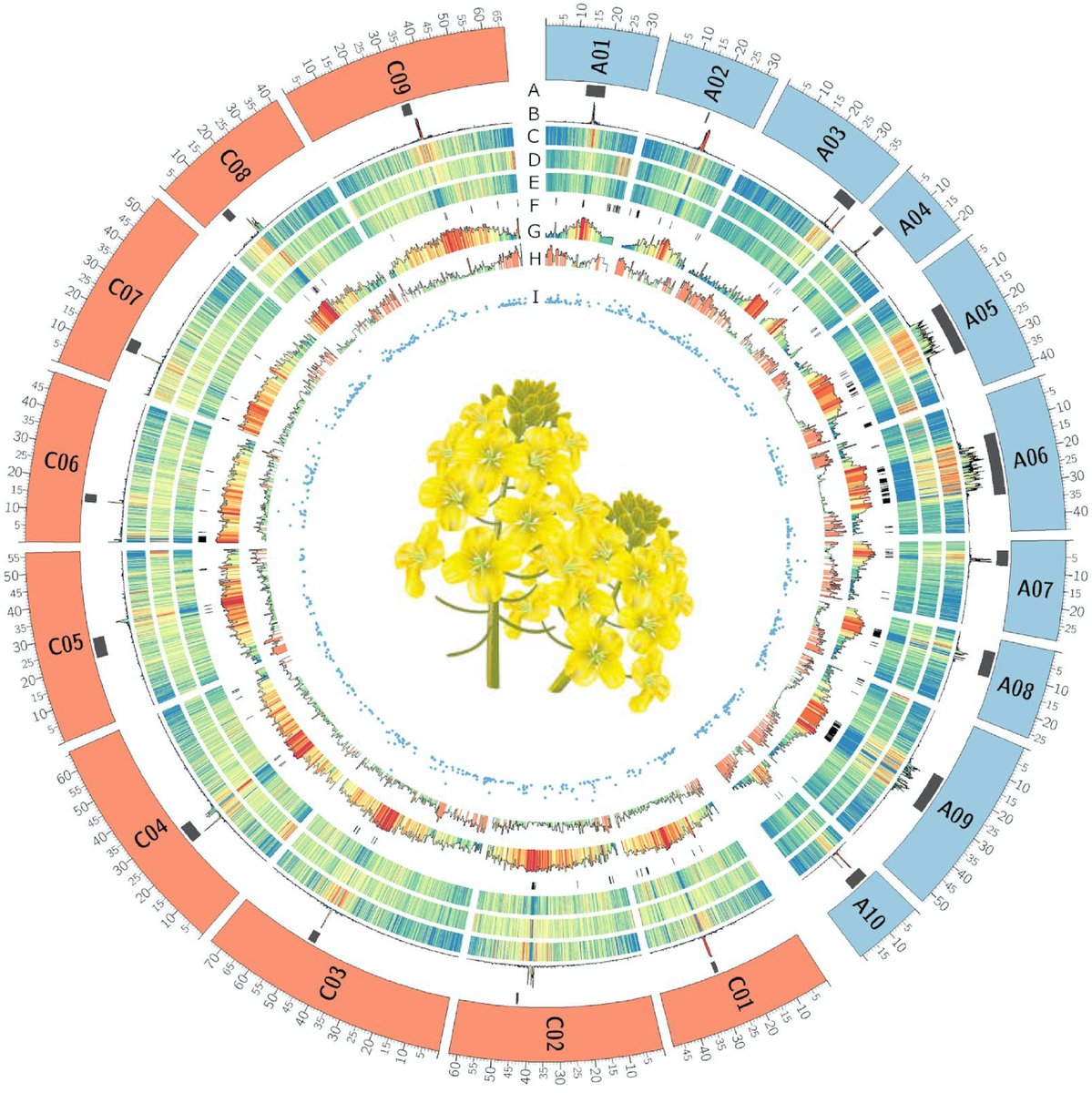
Long-read (@nanopore PromethION) assembly of the Brassica napus reference genome Darmor-bzh. Taking advantage of direct RNA sequencing to annotate the genome and focus on resistance genes doi.org/10.1093/gigasc…
Improving assemblies with optical maps peerj.com/articles/10150/ via @thePeerJ
the existing ONT interface (MinKnow) does not provide enough plots or interactivity to explore the quality of sequencing data in depth. BoardION dedicated to sequencing platforms, for real-time monitoring of all ONT sequencing devices; MinION, Mk1C, GridION and PromethION.
BoardION: real-time monitoring of Oxford Nanopore Technologies devices biorxiv.org/cgi/content/sh… #biorxiv_bioinfo
BoardION: real-time monitoring of Oxford Nanopore Technologies devices biorxiv.org/cgi/content/sh… #bioRxiv
@skimomiks mentioning Pore-C (long read concatemers for conformation capture) to resolve complex SVs @nanopore @WeillCornell #AGBT20

Live demo of the Mk1C MinION with @angelicavittor1 and @FrancoisBerdou during joint ONT/Agilent/NVIDIA seminar @Genomique_ENS #Genomics


latest progress! A total of 37 cells were sequenced using @nanopore PromethlON in a monocotyledon sequencing project, yielding 1282.7Gb ultra-long reads! The average N50 is 51.9Kb, and a single flow cell reads N50 is up to 143.3Kb! Expect the genome assembly of ultra-long reads!

Main takeaways on getting the best performance from guppy. - when using the client/server mode partition data and use multiple clients - use nvidia-smi to check the power draw is high on each gpu - use fast storage/networking so I/O isn’t bottlenecking #nanoporenvidia

Guppy now does mod bases.
Comparative assessment of long-read error-correction software applied to RNA-sequencing data biorxiv.org/content/10.110…
🐜 @ynsect annonce une levée de fonds 110 millions d'€. L'entreprise élève des insectes et les transforme en ingrédients pour animaux. Ses laboratoires sont situés au @Genopole à @VilledEvryCourc ▶️ Regardez notre reportage réalisé en 2017 : youtu.be/Fkx-18panpg
Today at 10:30 in the Pacific Salon @J_M_Aury will discuss 'Chromosome-Scale Assemblies of Wild Musa Genomes using Nanopore Long Reads and Optical Maps' #PAGXXVII #PAG2019

Ahead of @J_M_Aury's talk at the #PAGXXVII workshop read his Nature publication 'Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps': bit.ly/2RmpxqQ #PAG2019

Minimap2 v2.15 released with a few bug fixes. Available via github, bioconda and pypi (for mappy). This version outputs alignments identical to v2.14. github.com/lh3/minimap2/r…
MinION Mk 1c will combine MinION, MinIT for rapid data analysis and a screen, for a one-stop palm sized fully portable sequencing system, fully connected. Targeting mid 2019 for release but we'll open the shop in January. #nanoporeconf

New flappie basecaller showing some solid improvements on both amplicon and native DNA. ~3% point bump on individual read, 2.8x speed hit. @nanopore

United States Trends
- 1. ICBM 97,2 B posts
- 2. Dnipro 31,7 B posts
- 3. Good Thursday 21,4 B posts
- 4. #ThursdayMotivation 3.964 posts
- 5. Friday Eve N/A
- 6. #21Nov 1.907 posts
- 7. $DUB 6.727 posts
- 8. #thursdayvibes 2.641 posts
- 9. #Wordle1251 N/A
- 10. Nikki Haley 27,2 B posts
- 11. #KashOnly 1.440 posts
- 12. Adani 656 B posts
- 13. Bitcoin 640 B posts
- 14. Happy Birthday Nerissa 7.258 posts
- 15. Juice WRLD 23 B posts
- 16. Thomas Sowell 9.614 posts
- 17. Ellen DeGeneres 67,2 B posts
- 18. Paul George 9.494 posts
- 19. SWARM 7.361 posts
- 20. Jimmy Fallon 102 B posts
Who to follow
-
 France Génomique
France Génomique
@fr_genomics -
 Bioinfo Genotoul
Bioinfo Genotoul
@BioinfoGenotoul -
 David Vallenet
David Vallenet
@vallenet -
 Jean-Marc Aury
Jean-Marc Aury
@J_M_Aury -
 GeT_Genotoul
GeT_Genotoul
@GeT_Genotoul -
 INRAE Genomics
INRAE Genomics
@INRAE_Genomics -
 LAGE Genoscope
LAGE Genoscope
@GenoLAGE -
 LABGeM
LABGeM
@GenoLabgem -
 Guillaume GAUTREAU
Guillaume GAUTREAU
@GUILLAUMEGAUTRE -
 EPGV
EPGV
@EPGV_INRAE -
 Pedro H Oliveira
Pedro H Oliveira
@pholive81 -
 Kévin Lebrigand, PhD
Kévin Lebrigand, PhD
@kevinlebrigand -
 Chloé Baum
Chloé Baum
@ch_baum -
 Olivier
Olivier
@olivierlucas29 -
 Alexis Groppi
Alexis Groppi
@agroppi
Something went wrong.
Something went wrong.