Kristina Gomoryova
@kristina_gomoryResearcher at @RECETOX, Brno. Mass spectrometry, metabolomics, proteomics, R, data analysis.
Similar User

@Jakub_Harnos

@wilhelm_compms

@StoyanStoychev1

@MarketaKau

@souceklab

@bryjalab

@AJ_Brenes

@KrivanekLab

@AntoniaMikulova

@t_barta

@michaelsteidel1

@MichaelaNoskov5

@TRadaszkiewicz

@jspaezp1

@Kohoutkova_El
RNAseq is commonly performed. We compared N=30 wild-type Bl6 mice to an N=30 heterozygotic knockout with a phenotype, and then downsized. We found you need at least an N= 8-12 to avoid a >50% false positive rate. A 2nd line confirmed the finding. biorxiv.org/content/10.110…
Our paper describing a new method (FLiP-MS) for the global profiling of protein complex dynamics is out @NatureBiotech ! Read here👉: doi.org/10.1038/s41587… 🔬 Highlights: - Uses an experimental library of peptide markers integrated with a structural proteomics workflow. -…
We report many proteins not predicted by the genetic code. They are stable & abundant O( 10³ ) copies / cell. Generative mechanisms include codon-anticodon mismatches & RNA modifications. Their abundance depends on codon frequency & protein stability. biorxiv.org/content/10.110…
Happy to see #quantms out nature.com/articles/s4159…. Thanks to the main authors Dai & Julianus. The @OpenMSTeam @OKohlbacher & @DemichevLab for the support with OpenMS and DIANN. Thanks to @lukas_k for his incredible work with the data. Thanks to @nf_core & @nextflowio platforms.
A long awaited and important paper is finally out: pubs.acs.org/doi/10.1021/ja…. In a classic 🇸🇪 math textbook there is a cartoon that says "löst värde - värdelöst", roughly translated as "loose value - worthless". Like mass spectrometry data without QC or uncertainty.
This computational mass spec training fellowship sounds like an awesome opportunity I would have liked to see as a grad student or a postdoc back in those days..
We are happy to support the @OpenMSTeam fully funded fellowship training in computational MS! 🤗 Great opportunity for graduate students and recent grads 👩🎓 Scientists from underrepresented backgrounds are especially encouraged to apply. More info 👉openms.org/news/fellowshi…

Did you know that 28 out of 34 signalling pathways, and ~60% of the proteome, respond differently depending on the metal ion environment? Neither did we, but now believe that the metal ion levels are one of the big "known unknowns" in cellular networks biorxiv.org/content/10.110…
rworkflows: automating reproducible practices for the R community | Nature Communications nature.com/articles/s4146… #Bioinformatics



A free package for learning and practicing gene set enrichment analysis jokergoo.github.io/GSEAtraining/

I think the sad actual answer to "What is the biggest open challenge in biology?" is: 1. Getting people to share data. 2. Structuring, organizing, and annotating data with metadata so it's useful. 3. Building higher-level abstractions so people can efficiently work with big data.
What are you favorite methods for building biological networks from omics data? @AcademicChatter #AcademicTwitter
Our new preprint is available on biorXiv: doi.org/10.1101/2023.1… !🥳 We present scplainer, a principled computational approach to streamline and standardise #MassSpec #SingleCell #proteomics data analysis. Here's a 🧵 with our key messages.

🚨 It's here 🚨 The most comprehensive overview of modern proteomics ever written. ▶️ 19 figures ▶️ 17 chapters ▶️ 111 pages ▶️ 859 references Contributions from: ▶️ 19 authors ▶️ 13 institutions ▶️ 6 countries ▶️ 4 continents arxiv.org/abs/2311.07791
Having started proteomics 2 yrs ago now, me and my fabulous colleagues (@csdawsonn, @lisabreckels & @CoralTKrueger) put together a workflow containing all of the things I wish that I had known about analysing expression proteomics experiments. f1000research.com/articles/12-14…
the more i learn about statistics the more horrified i get at how much personal interpretive choice is at play at every step of the process. i came in expecting a hard science and feel like i'm getting slapped with way more art than i expected
1/ This is what happens when you give 132 teams of researchers the SAME DATASET and ask the SAME QUESTION: How does sibling competition affect nestling growth in blue tits? Most teams found negative effect, but large variation in effect size. Some teams no or positive effect

Straight from the cookbook of p-hacking: “Add a twist of surprise to the paper”
No no no no no no no no no. Thankfully, this advise was ignored by the authors. But this wide spread but unspoken belief is why NeurIPS/ICML/ICLR reviewing for empirical papers is totally broken.

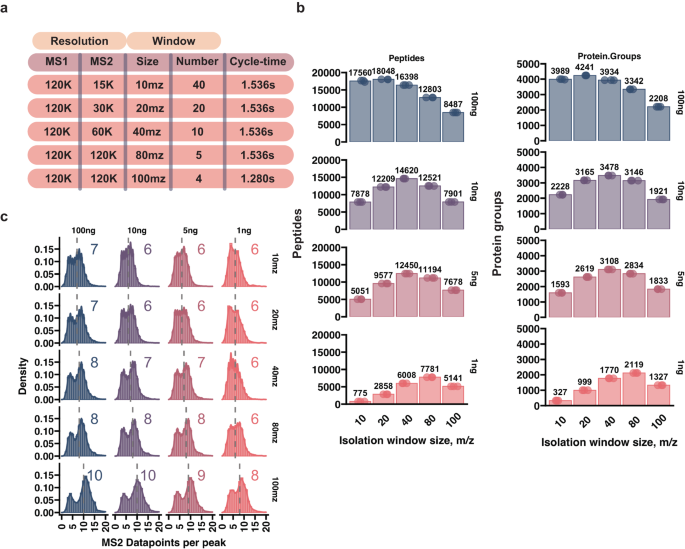
@NotValdemaras from @erwinschoof lab and colleagues leverage sensitivity-tailored data acquisition methods for scpMS to profile cell state heterogeneity in cultured model systems nature.com/articles/s4146…
Paper alert! Here is our fast and easy protocol to reduce strong signals of ligand peptides from nanobody peptides or streptavidin in AP-MS/BioID experiments. Collaboration between @MaxPerutzLabs , @StAnna_CCRI & @KraftLabFR A tweetorial!⬇️ pubs.acs.org/doi/10.1021/ac…
Introducing DROPPS! Here's our preprint that describes an accesible approach to low input proteomics and how we applied it to generate some interesting biological insights (1/x) biorxiv.org/content/10.110…
United States Trends
- 1. Justin Tucker 12,5 B posts
- 2. Steelers 40,1 B posts
- 3. Drake Maye 5.246 posts
- 4. $CUTO 8.771 posts
- 5. Packers 21,8 B posts
- 6. #HereWeGo 6.650 posts
- 7. #RavensFlock 3.647 posts
- 8. Jordan Love 3.249 posts
- 9. McDonald 69,3 B posts
- 10. Browns 16,1 B posts
- 11. Taysom Hill 1.500 posts
- 12. #GoPackGo 3.157 posts
- 13. #Jets 2.995 posts
- 14. #BALvsPIT 2.341 posts
- 15. Gibbs 5.576 posts
- 16. Clark Kent 3.635 posts
- 17. Coke 42,8 B posts
- 18. DeFi 102 B posts
- 19. Najee 1.561 posts
- 20. Sauce Gardner N/A
Who to follow
-
 Harnos_Lab
Harnos_Lab
@Jakub_Harnos -
 Mathias Wilhelm
Mathias Wilhelm
@wilhelm_compms -
 Stoyan Stoychev
Stoyan Stoychev
@StoyanStoychev1 -
 Kaucka Marketa
Kaucka Marketa
@MarketaKau -
 SoucekLab
SoucekLab
@souceklab -
 Bryjalab
Bryjalab
@bryjalab -
 Alejandro Brenes
Alejandro Brenes
@AJ_Brenes -
 Jan Krivanek
Jan Krivanek
@KrivanekLab -
 Antónia Mikulová
Antónia Mikulová
@AntoniaMikulova -
 Tomas Barta
Tomas Barta
@t_barta -
 Michael Steidel
Michael Steidel
@michaelsteidel1 -
 Michaela Noskova Fairley
Michaela Noskova Fairley
@MichaelaNoskov5 -
 Tomasz Radaszkiewicz
Tomasz Radaszkiewicz
@TRadaszkiewicz -
 Sebastian Paez
Sebastian Paez
@jspaezp1 -
 Eliška Kohoutková
Eliška Kohoutková
@Kohoutkova_El
Something went wrong.
Something went wrong.