Ian Dunn
@ian_dunn_PhD Candidate @CMUPittCompBio, advised by @david_koes. Interested in deep learning for molecular structures and accelerating drug discovery.
Similar User

@MirunaCretu2

@OSchilter

@WenhaoGao1

@DerekvTilborg

@6ojaHa

@drugilsberg

@lucky_pattanaik

@BaillifBen

@YuanqingWang

@JeffGuo__

@MorganThomas263

@JannisBorn

@Ye_Buehler

@AdeeshKolluru

@DavidKreutter
New preprint! “Mixed Continuous and Categorical Flow Matching for 3D De Novo Molecule Generation” arxiv.org/abs/2404.19739… FlowMol is a flow matching model that jointly samples the geometric and topological structure of molecules. We also present surprising results on flow…
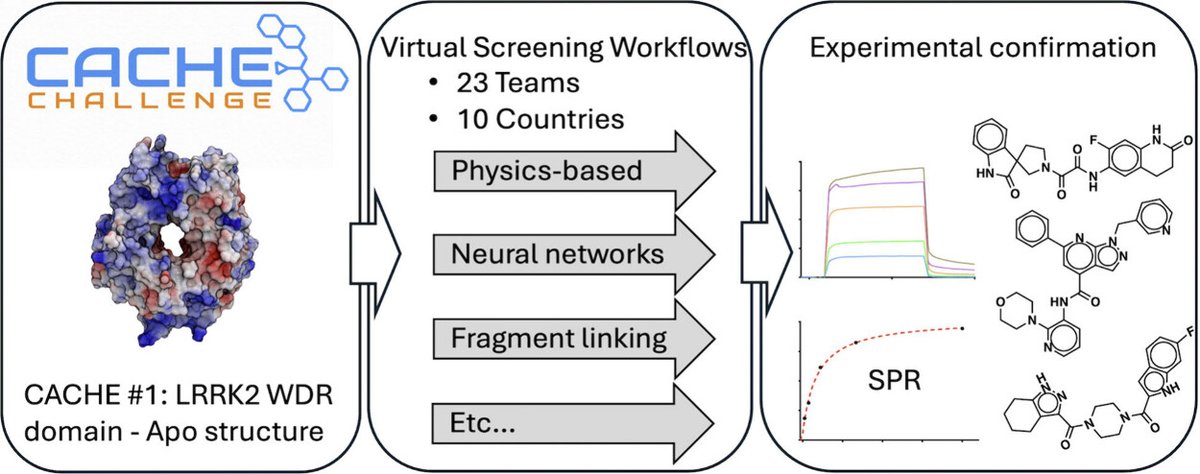
Finally, a community-wide paper @JCIM_JCTC led by @mattschap describing results of CACHE Challenge #1: Targeting the WDR Domain of LRRK2, A Parkinson’s Disease Associated Protein! #compchem #drugdiscovery pubs.acs.org/doi/10.1021/ac… Here we report the results of the inaugural…
Our work on all-atom tokenization of any biomolecular structure is on arxiv now! arxiv.org/abs/2410.19110 @BiologyAIDaily summary is perfect, but to sum up the summary: - composability: small molecule, protein and RNA structure "point-clouds" can be described with the same vocab…
Bio2Token: All-atom tokenization of any biomolecular structure with Mamba @FlagshipPioneer • This paper introduces “Bio2Token”, a method that tokenizes biomolecular structures at an all-atom level using Mamba. Unlike many current approaches that rely on coarse-grained…

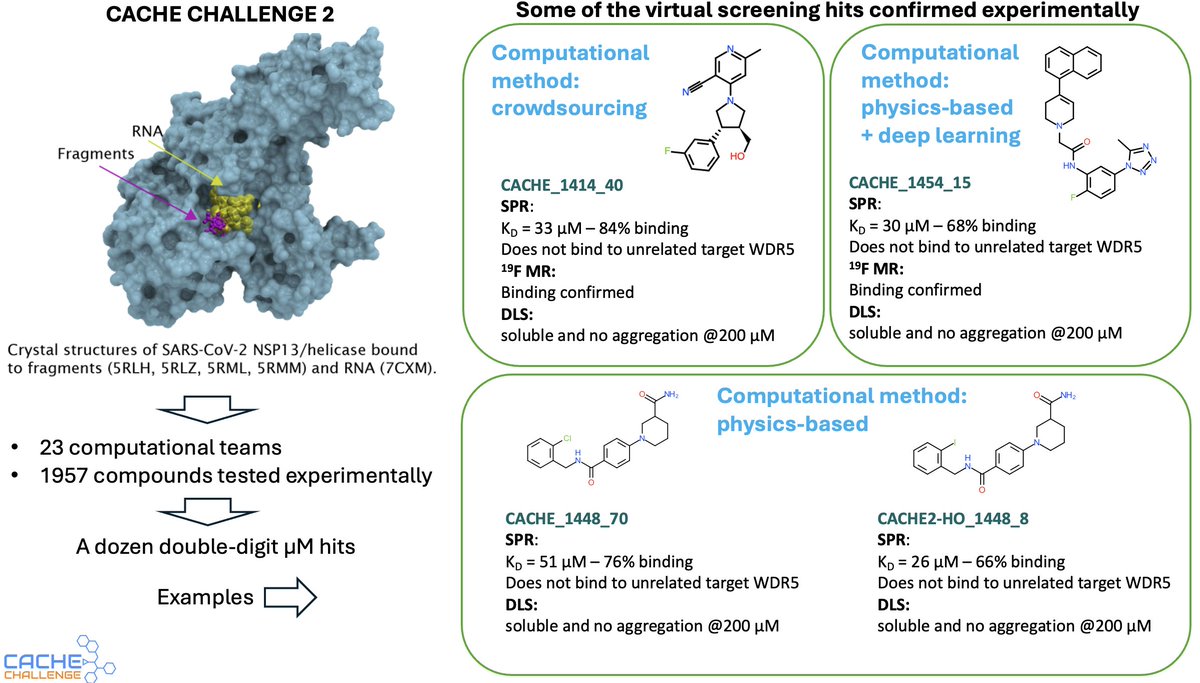
Announcing CACHE2 results! Crowdsourcing and conventional methods did as well as novel ML. Teams started from 4 fragments in the PDB. We tested their predictions and found double-digit µM hits. This is the state-of-the-art. @thesgconline @consciencemeds bit.ly/4dTPBzO
We’re presenting AlphaProteo: an AI system for designing novel proteins that bind more successfully to target molecules. 🧬 It could help scientists better understand how biological systems function, save time in research, advance drug design and more. 🧵 dpmd.ai/3XuMqbX
ESM3 is out from EvolutionaryScale! 98B params (~GPT3 scale). Multimodal over sequence, structure, and function with cool design applications. Trained on variable masking ratios and decodes proteins iteratively. Some work on alignment too. Looks exciting! evolutionaryscale.ai/blog/esm3-rele…
Great post on AlphaFold3 from @inductive_bio that I'm surprised has not got more attention🤔 - AF3 paper benchmarked Vina (not SOTA) using a single conf - Conventional docking has near AF3 performance when using multiple starting confs and Gnina scoring to select best pose (1/2)

Excited to share our latest preprint: "Transferable Boltzmann Generators"! We propose a framework based on flow matching and demonstrate transferability on dipeptides. Work done with amazing @FrankNoeBerlin Check it out here: arxiv.org/abs/2406.14426
New blog post alert! 🚨 I write about AlphaFold3, how it works, where it works well ✅ (and where we think it might not ❌) and what it means to the TechBio landscape more broadly (if you decide to believe me). 🧬 (1/7) 🧵 open.substack.com/pub/harrisbio/…

Introducing FoldFlow-2, a new SOTA sequence-conditioned protein generative model! work w/ @DreamFoldAI James V @FatrasKilian Eric TL @PabloLemosP @riashatislam @ChengHaoLiu1 @jarridrb @tara_aksa @mmbronstein @AlexanderTong7 @bose_joey Arxiv: arxiv.org/abs/2405.20313 1/8 🧵
Cool new paper with Julian Cremer, Tuan Le, Okko Clevert and @ktschuett, introducing a new way to do conditional generation through diffusion models via importance sampling of the denoising pathway. Applied to ligand generation in proteins. arxiv.org/abs/2405.14925
Wonder how to generate discrete data with Riemannian Flow Matching? Well, here is our most recent work addressing this issue: Fisher Flow Matching 😎 ➡️ Available here: arxiv.org/abs/2405.14664 🔥 Let me break it down 🧵 (1/10) @bose_joey @mmbronstein @ismaililkanc @MirceaSci

Wonderful critical evaluation of a generative model for SBDD. Bad news: generated small molecules require heavy post-processing and many compounds are unstable. Good news: This provides clear goal posts for model performance that we should be working towards.
New Practical Cheminformatics Post, "Generative Molecular Design Isn't As Easy As People Make It Look", practicalcheminformatics.blogspot.com/2024/05/genera…

I am happy to share our new ICML work, a new score-based generative model for molecules conditioned on protein structures. arxiv.org/abs/2405.03961 (below is an example of a single-chain generating ligands)
This raises many questions. Did anybody ever compare the performance of frame representations and equivariant architectures to simpler options? I think I’ve seen results suggesting equivariance helps. Why have researchers come to different conclusions about the utility of…
AF2: inspires a zillion diffusion/flow matching papers to use a frame representation of the backbone, AF3: diffusion over atomic coordinates with no equivariances built into the architecture or representation.
Very excited to present SynFlowNet next week at @gembioworkshop #ICLR2024! We tackle the synthesisability issue in molecular design by training a GFlowNet with an action space of chemical reactions. Thanks to my coauthors @charlieharris01 @juleroy13 @folinoid and Pietro Lio (1/n)
We're excited to have @ian_dunn_ from the @david_koes come to the Rosetta Molecular ML Reading Group to present his work on "keypoint" latent molecular diffusion models Today at 11am EDT. See maomlab.github.io/mol_ml_reading… for the zoom link
David Koes and Ian Dunn: Docking with GNINA is all you need!

United States Trends
- 1. Ohio State 25 B posts
- 2. Indiana 35,7 B posts
- 3. $AROK 6.434 posts
- 4. Wayne 125 B posts
- 5. Howard 22,7 B posts
- 6. #daddychill 2.389 posts
- 7. Ryan Day 3.740 posts
- 8. Gus Johnson N/A
- 9. Buckeyes 7.108 posts
- 10. Hoosiers 7.479 posts
- 11. #iufb 4.326 posts
- 12. $MOOCAT 7.665 posts
- 13. DJ Lagway 1.751 posts
- 14. UMass 3.061 posts
- 15. Tottenham 41,8 B posts
- 16. Surgeon General 113 B posts
- 17. Chip Kelly N/A
- 18. Man City 39,2 B posts
- 19. Carnell Tate N/A
- 20. Cody Simon N/A
Who to follow
-
 Miruna Cretu
Miruna Cretu
@MirunaCretu2 -
 Oliver Schilter
Oliver Schilter
@OSchilter -
 Wenhao Gao
Wenhao Gao
@WenhaoGao1 -
 Derek van Tilborg
Derek van Tilborg
@DerekvTilborg -
 BOjana Ranković
BOjana Ranković
@6ojaHa -
 Matteo Manica
Matteo Manica
@drugilsberg -
 Lucky Pattanaik
Lucky Pattanaik
@lucky_pattanaik -
 Benoît Baillif
Benoît Baillif
@BaillifBen -
 Yuanqing Wang
Yuanqing Wang
@YuanqingWang -
 Jeff Guo
Jeff Guo
@JeffGuo__ -
 Morgan Thomas
Morgan Thomas
@MorganThomas263 -
 Jannis Born
Jannis Born
@JannisBorn -
 Ye Buehler
Ye Buehler
@Ye_Buehler -
 Adeesh Kolluru
Adeesh Kolluru
@AdeeshKolluru -
 David Kreutter
David Kreutter
@DavidKreutter
Something went wrong.
Something went wrong.