Similar User

@juhyunk_

@SCAID__

@Seyoon_L

@AquilasRecruit

@cestlevy

@Healthcareonli2

@verpetrova

@EamonFaulkner98

@ElsallabMagdi

@WestWiktor

@Scientist_Melb
Genome engineering excelled at small nucleotide changes, but large edits remained challenging. Recent innovations are changing this. We reviewed the field of structural variant engineering @NatureGenet 🧬 @JulianeWeller @TVanderstichele @LeopoldParts nature.com/articles/s4158…
Thrilled to share our paper published in @Nature, where we uncover a new process of tumor initiation through a polyclonal-to-monoclonal transition at the @precancerous stage. So proud of my fantastic team (led by @ZhaolianLu ) and incredible collaborators! nature.com/articles/s4158…

A paper in @Nature describes a new method for DNA-based data storage that may enhance the speed and cost-effectiveness of writing data into DNA. go.nature.com/4hh9QJG

Excited to report our study in @NEJM on the discovery of deletions in a long noncoding RNA gene 🧬 (𝘊𝘏𝘈𝘚𝘌𝘙𝘙) as the cause of a newly defined human neurodevelopmental disorder 🧠. 🧵1/10 nejm.org/doi/full/10.10…
In @Nature we use single cell multiomics, fibroblast fate tracing, and in vivo models to characterize immune fibroblast crosstalk in heart failure with @LavineLab @washumedicine, @Amgen , and @ImmunoFibHF nature.com/articles/s4158…
It's great to see my first paper of the PhD published in @NatureBiotech! go.nature.com/47ARizC. This describes our scRNA-seq method, scITD, for analyzing how the transcriptional states of multiple cell types are linked across patients. A few highlights from the paper:
1/🧵Excited to re-introduce Memento in (cell.com/cell/fulltext/…), a scalable method for differential analysis of scRNA-seq data. It provides first-in-class performance in statistical power and calibration for comparing differences in mean, variability, and gene correlation.👇
How the small intestine is spatiotemporally regulated by its unique nutritional environment @CellCellPress cell.com/cell/abstract/…
A just-published paper in Nature Biotechnology from the lab of C4E Director Bing Ren describes a novel technique, Droplet Hi-C, for profiling chromatin both more effectively and at a lower cost. The paper also sheds new light on extra-chromosomal DNA.
Droplet Hi-C enables scalable, single-cell profiling of chromatin architecture in heterogeneous tissues go.nature.com/3NCqwO1

Interpretable multi-omics integration with UMAP embeddings and density-based clustering biorxiv.org/content/10.110…
Statistically principled feature selection for single cell transcriptomics biorxiv.org/cgi/content/sh… #biorxiv_bioinfo
Check out our latest #Roadmap: Olga Anczukow @OlgaAnczukow @jacksonlab, Andrei Thomas-Tikhonenko @andrei_thomas_t @PennMedicine & Co explore advances in #splicing biology and provide guidance on leveraging these insights to target splicing in cancer. go.nature.com/3YrvkfH
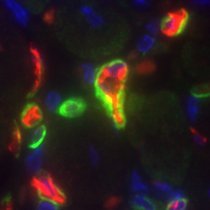
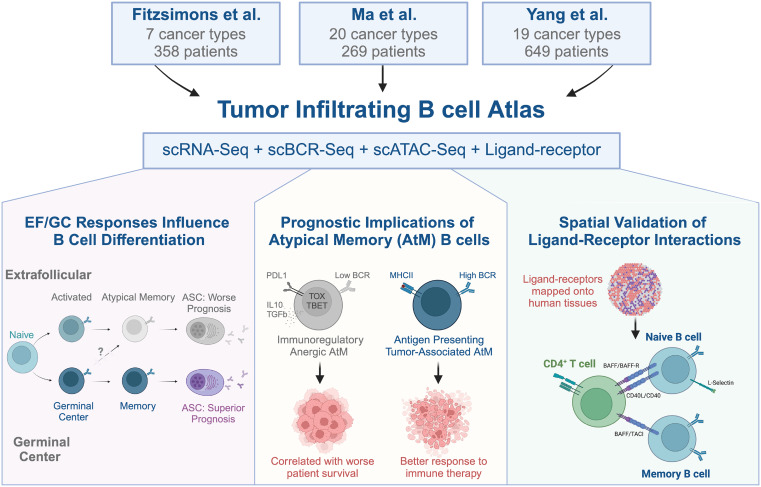
ScRNA-seq defines pan-cancer atlas of tumor-infiltrating B cell subsets Preview @Cancer_Cell cell.com/cancer-cell/fu… cell.com/cancer-cell/fu… doi.org/10.1016/j.cell… science.org/doi/10.1126/sc…
NEW content online! Epigenomic heterogeneity as a source of tumour evolution go.nature.com/4dKLSUg

A pan-cancer single-cell RNA-seq atlas of intratumoral B cells dlvr.it/TFNVLL

🚀 We're thrilled to introduce Orthrus 🧬🐕—a groundbreaking mature RNA foundation model designed to push the boundaries of RNA property prediction! 🔬 What is Orthrus? Orthrus is a Mamba-based RNA foundation model, pre-trained using a novel self-supervised contrastive learning…
Analysis of single-cell CRISPR perturbations indicates that enhancers predominantly act multiplicatively dlvr.it/TFL7y4
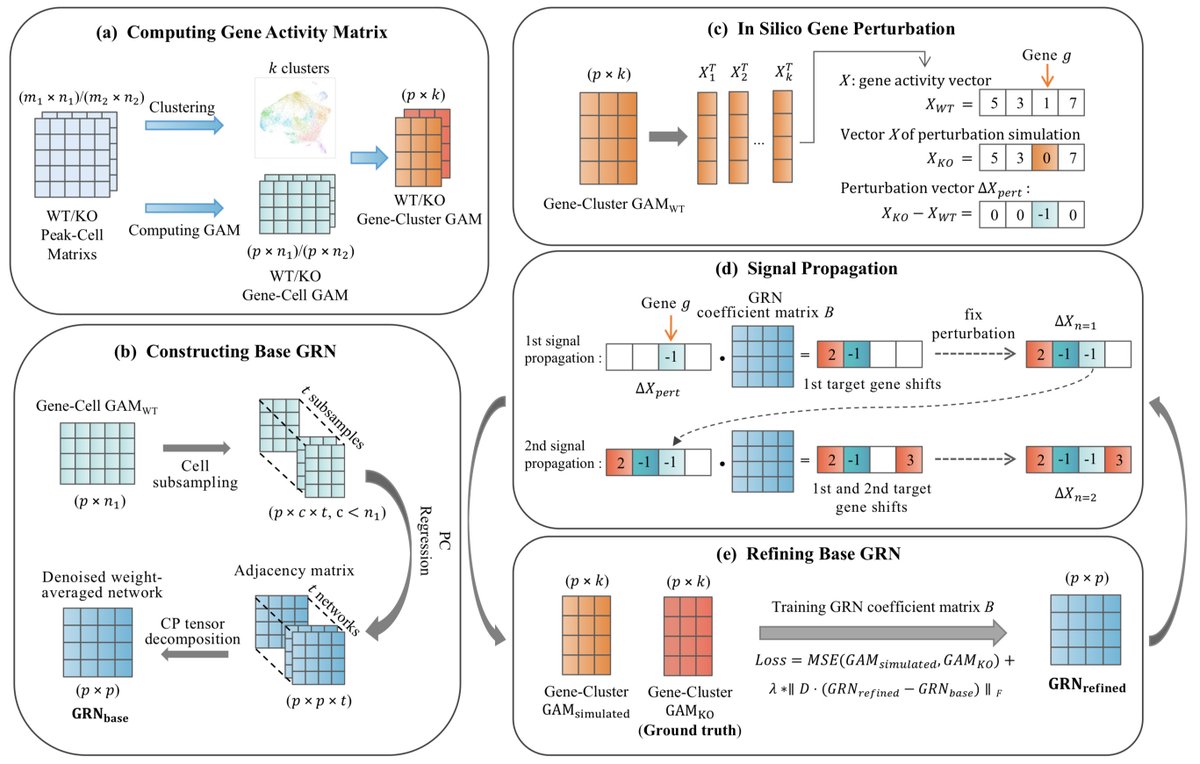
Inferring Gene Regulatory Network Based on scATAC-seq Data with Gene Perturbation biorxiv.org/content/10.110…
Defining precancer: a grand challenge for the cancer community. #Precancer @NatureRevCancer nature.com/articles/s4156…
United States Trends
- 1. #UFCMacau 21,8 B posts
- 2. #ArcaneSeason2 172 B posts
- 3. Wayne 93,6 B posts
- 4. Leicester 38,5 B posts
- 5. Good Saturday 24,5 B posts
- 6. Jayce 72,6 B posts
- 7. Ekko 70,9 B posts
- 8. #saturdaymorning 3.290 posts
- 9. Nicolas Jackson 7.345 posts
- 10. #LEICHE 19,9 B posts
- 11. Ndidi 3.057 posts
- 12. Nico Jackson 1.224 posts
- 13. Wang Cong 1.391 posts
- 14. SEVENTEEN 1,43 Mn posts
- 15. Gabriella Fernandes N/A
- 16. #SaturdayVibes 4.715 posts
- 17. Sanchez 228 B posts
- 18. Jinx 199 B posts
- 19. maddie 22 B posts
- 20. Ricci 2.265 posts
Who to follow
Something went wrong.
Something went wrong.