Aleix Lafita
@alafitamasipComputational proteinologist @GSK . Views are my own. @.alafita.bsky.social
Similar User

@philipp_walch
@AI_Proteins

@bahl_lab

@ProteinBoston

@denbiOffice

@MatteTrovato23

@Structjon

@Alexbateman1

@SBGrid

@pcabezaspadilla

@KingLabIPD

@EMBLEBIjobs

@cgbarker_

@Dey_Gautam

@PPMC_UAB
"Fine-tuning Protein Language Models with Deep Mutational Scanning improves Variant Effect Prediction" 📢 We have uploaded the final camera-ready version of the paper that @ferrangh and I will present at the @MLGenX workshop at #ICLR2024 this May openreview.net/pdf?id=d7AdTpY… 🧵1/5

With David and the Baker Lab in the spotlight today, I wanted to share some insights into the @UWproteindesign and how it operates, a glimpse behind the curtain. I had planned to write this post-graduation, but now seems as good a time as any. (Got twitter blue free trial so this…
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”

The genetic architecture of protein stability by @aj4re @ainamartiaranda @cristonah @ToniBeltran13 @JoernSchmiedel nature.com/articles/s4158… @CRGenomica @sangerinstitute @ALLOXbio
Exciting new work from Qian Cong's group on predicting human protein interactome. Leveraging new eukaryotic genomes, new RoseTTAFold2 trained on +/- pairs of PPI and large distilled dataset of domain-domain interactions! 🤩 biorxiv.org/content/10.110…

Our latest from @TheCrick, now on @biorxivpreprint: biorxiv.org/content/10.110… Michael Herger (@HergerMichael) and Christina Kajba jointly led development of a new prime editing platform for testing large numbers of human genetic variants.
Our work led by @megan_buckley01 on Saturation Genome Editing of VHL is now out in @NatureGenet: nature.com/articles/s4158… Many new experiments and analyses to quickly highlight… (1/n)
Weekend project: Comparing ESM3 from @EvoscaleAI to ESM2 and inv_cov. The ultimate test of a protein language models is how well the pairwise dependencies it learns correlate to structure. (1/8)

We have trained ESM3 and we're excited to introduce EvolutionaryScale. ESM3 is a generative language model for programming biology. In experiments, we found ESM3 can simulate 500M years of evolution to generate new fluorescent proteins. Read more: evolutionaryscale.ai/blog/esm3-rele…
ProtMamba: a long-context protein language model trained by concatenating homologous sequences. @damiano_sga @CMalbranke

Today in @Nature we present the results of our exploration of the soluble protein universe and the design of functional soluble membrane proteins with @CasperGoverde @GoldbachNico @befcorreia 1/6 nature.com/articles/s4158…
In the latest edition of "Huh, I didn't expect that to work", our latest paper shows that LLMs can outperform existing methods for identifying causal genes in genome-wide association studies. 🧵 medrxiv.org/content/10.110…
Great opportunity, the job post is still open!
R-U-seeking a postdoc at the interface of functionalGenetics, targetDiscovery, CRISPR-data and #AI? liked these methods: shorturl.at/jlrD2 rb.gy/xv04n8 rb.gy/y7ydpj rb.gy/rjf6hlwant beyond cancer? JOIN US! @humantechnopole @OpenTargets plsRT🙏

The protein design & informatics group at @GSK is looking to hire a computational biologist in the UK and/or switzerland jobs.gsk.com/en-us/jobs/384…
The end goal is not to replace variant effect predictors (like AlphaMissense, EMS or EVE) but to provide a predicted mechanism of effect to overlay on these predictors. For example, cancer hotspot mutations tend to have a higher fraction of variants at interfaces than clinvar.

Great preprint and resource from @OpenTargets with contributions from our team at GSK!
New preprint: sequence-based models are very good at predicting human protein missense variants with an impact on function. Here we use AlphaFold2 models to infer mechanisms of effect of deleterious variants (stability, pockets and protein interfaces) biorxiv.org/content/10.110…
Introducing Genie 2, our latest protein design model! Genie 2 sets a new state of the art on key design metrics and supports motif scaffolding, including that of multiple motifs with undefined geometric relationships, a feature new to protein diffusion models. (1/5)
To help scientists explore whether a genetic variant is likely pathogenic, and which regions of proteins are functionally important, we've integrated AlphaMissense data by @GoogleDeepMind into @ensembl, @uniprot & the AlphaFold database. ebi.ac.uk/about/news/tec…

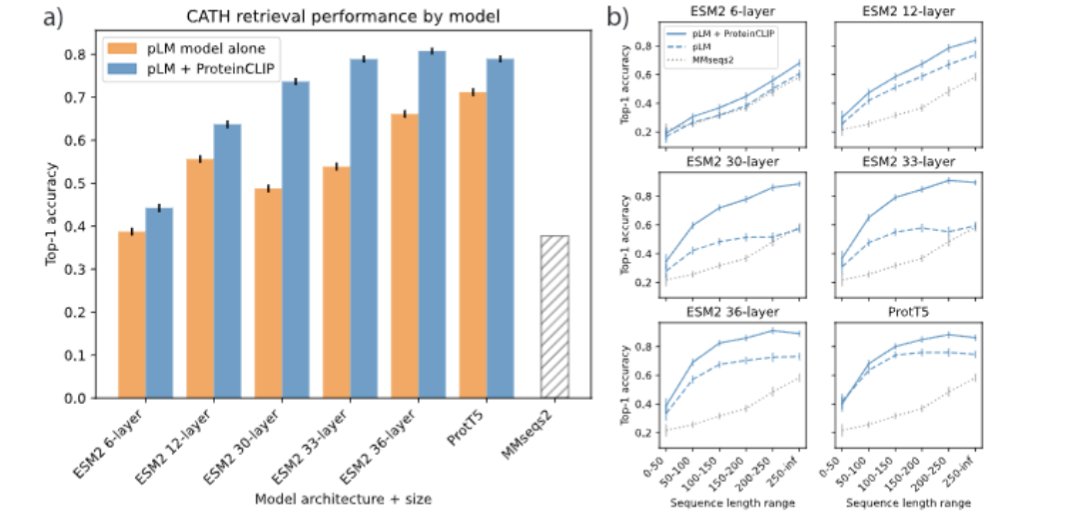
A CLIP-style contrastive model between protein sequences and text descriptions of their function. @Kevin_E_Wu @james_y_zou


new lab preprint - using proteomics and protein co-variation to infer protein association networks across 11 human tissues to study what determines tissue differences and to prioritize disease genes in GWAS linked genes through tissue specific networks biorxiv.org/content/10.110…
This is a great opportunity!
We have an open postdoc position in the area of protein structural bioinformatics. Areas of interest include proteome-wide models, variant effect prediction, molecular evolution, allosteric regulation, protein/drug design. Deadline 20th July jobs.ethz.ch/job/view/JOPG_…
We have an open postdoc position in the area of protein structural bioinformatics. Areas of interest include proteome-wide models, variant effect prediction, molecular evolution, allosteric regulation, protein/drug design. Deadline 20th July jobs.ethz.ch/job/view/JOPG_…
United States Trends
- 1. Colorado 52,6 B posts
- 2. Kansas 27 B posts
- 3. Ole Miss 31,9 B posts
- 4. Devin Neal 2.694 posts
- 5. Travis Hunter 9.581 posts
- 6. Indiana 62 B posts
- 7. Gators 18,8 B posts
- 8. Jaxson Dart 6.881 posts
- 9. Penn State 7.974 posts
- 10. Ewers 2.064 posts
- 11. Shedeur 7.618 posts
- 12. Shedeur 7.618 posts
- 13. Ohio State 40,8 B posts
- 14. Olivia Miles 1.729 posts
- 15. Sark 2.906 posts
- 16. Wayne 141 B posts
- 17. Heisman 7.683 posts
- 18. Fickell N/A
- 19. Minnesota 16,8 B posts
- 20. Nissan 23,6 B posts
Who to follow
-
 Philipp Walch
Philipp Walch
@philipp_walch -
 AI Proteins
AI Proteins
@AI_Proteins -
 chris bahl
chris bahl
@bahl_lab -
 Boston Protein Design and Modeling Club
Boston Protein Design and Modeling Club
@ProteinBoston -
 denbi
denbi
@denbiOffice -
 Matteo Trovato
Matteo Trovato
@MatteTrovato23 -
 Jonas Tholen
Jonas Tholen
@Structjon -
 Alex Bateman
Alex Bateman
@Alexbateman1 -
 SBGrid: Structural Biology Software
SBGrid: Structural Biology Software
@SBGrid -
 Patricia Cabezas
Patricia Cabezas
@pcabezaspadilla -
 King Lab
King Lab
@KingLabIPD -
 EMBL-EBI Jobs
EMBL-EBI Jobs
@EMBLEBIjobs -
 Charlie Barker
Charlie Barker
@cgbarker_ -
 Gautam Dey
Gautam Dey
@Dey_Gautam -
 Ventura's Lab
Ventura's Lab
@PPMC_UAB
Something went wrong.
Something went wrong.