Yuki Saito
@YukiSaito_MDPhDResearcher at @kataoka_lab and GI oncologist at @Keio_univ_PR. Views are my own. 国がん研究所/慶應消化器内科
Similar User

@kataoka_lab

@okada_yukinori

@TakashiKohno2

@friend1ws

@hikarundesuyo

@TMHemOnc

@NambaShinichi

@rmhtgy373south

@seiichi_mori

@nikuwoyakuman

@Know_the__Truth

@yota_ochi

@akiyama_mst

@changumu1

@HiromicsSuzuki
Happy to share our paper (my 1st paper as one of corresponding authors) out in @CD_AACR! Using >48,000 Japanese clinical sequencing data in #CCAT (@NccCat), we elucidated a pan-cancer landscape of driver alterations and their clinical actionability in Japanese patients. 1/3
Excited to share our latest paper from @CD_AACR We used >48,000 samples from C-CAT, a real-world clinico-genomic database in Japan, to elucidate driver alterations and their clinical actionability in Japanese patients. @Sara_Horie & @YukiSaito_MDPhD doi.org/10.1158/2159-8… (1/3)
Excited to share one of our apex studies from our @NCIHTAN center, focused on the transition to metastasis, shedding light transition to #CRC #metastasis. Joint work with @KarunaMDPhD, led by @andrewrmoorman and @elliebenitez11 --> nature.com/articles/s4158…
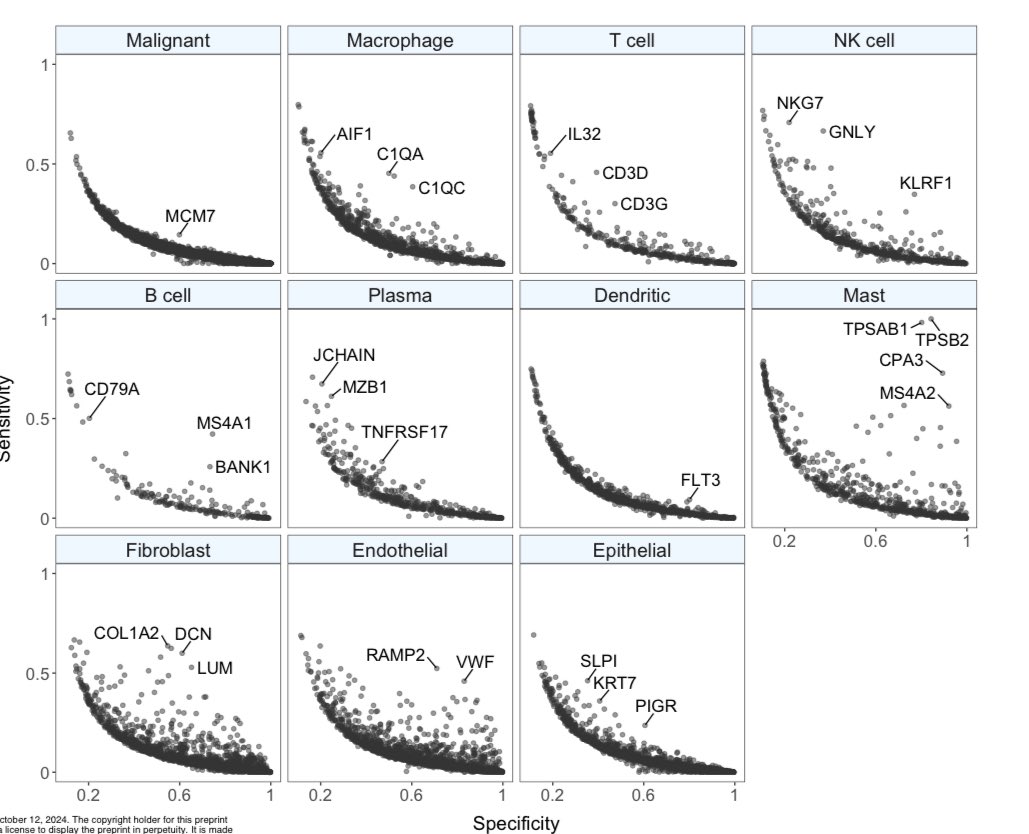
The @TiroshLab has another gem of a preprint. The Curated Cancer Cell Atlas: comprehensive characterisation of tumours at single-cell resolution biorxiv.org/content/10.110… ~125 datasets from across 40 cancer types, comprising ~2900 samples. 3CA website: weizmann.ac.il/sites/3CA/

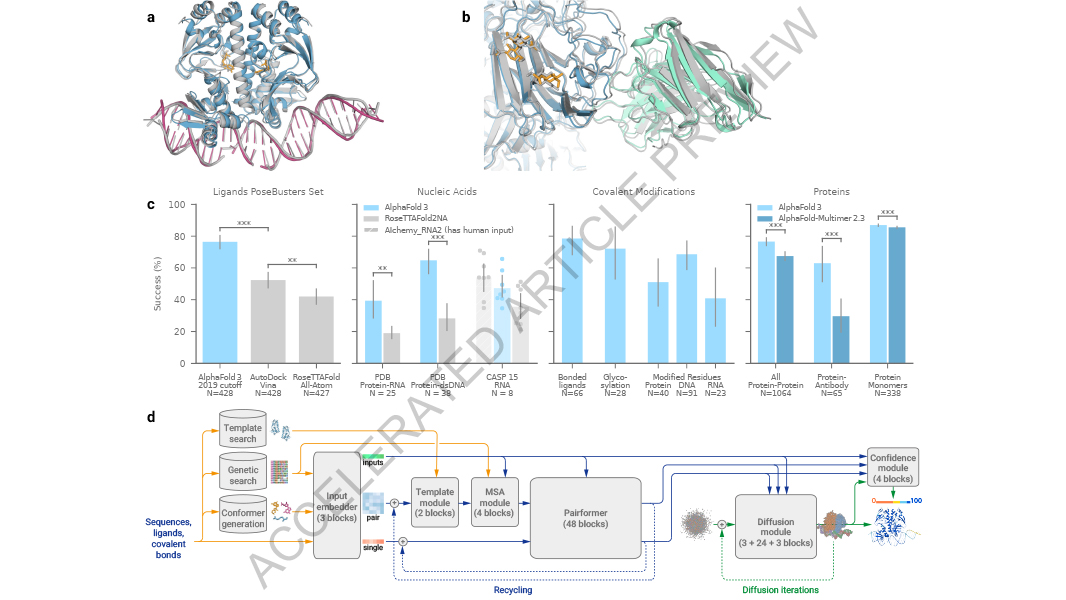
This year’s chemistry laureates Demis Hassabis and John Jumper have developed an AI model, AlphaFold2, to solve a 50-year-old problem: predicting proteins’ complex structures. Check out two examples of protein structures determined using AlphaFold2. First up, a bacterial enzyme…
The 2024 #NobelPrize laureates in chemistry Demis Hassabis and John Jumper have successfully utilised artificial intelligence to predict the structure of almost all known proteins. In 2020, Hassabis and Jumper presented an AI model called AlphaFold2. With its help, they have…

BREAKING NEWS The 2024 #NobelPrize in Physiology or Medicine has been awarded to Victor Ambros and Gary Ruvkun for the discovery of microRNA and its role in post-transcriptional gene regulation.

MHC Hammer reveals genetic and non-genetic HLA disruption in cancer evolution @CharlesSwanton @NickyMcGranahan and TRACERx team nature.com/articles/s4158…
Excited to share our latest manuscript in @NatureGenet! Utilizing high-resolution genetic data imputed with a population-matched reference panel, we uncovered causal genetic variants for a wide range of human traits over 200K participants from Japan (1/20) nature.com/articles/s4158…
9月26日、#国立がん研究センターは「国内初の造血器腫瘍 #遺伝子パネル検査「#ヘムサイト」の製造販売承認取得について」と題した記者会見を開催しました。 oncolo.jp/news/2401001ra…
Here we go! In Tokyo/Osaka Univ, we performed statistical fine-mapping of plasma protein expression QTLs (cis-pQTLs) in >1,000 Japanese individuals, for nearly 3,000 plasma protein expressions as measured by Olink. nature.com/articles/s4158… Seven highlights:
Approval of the first TCR-based cell therapy dlvr.it/TDWRtH #genetherapy #celltherapy
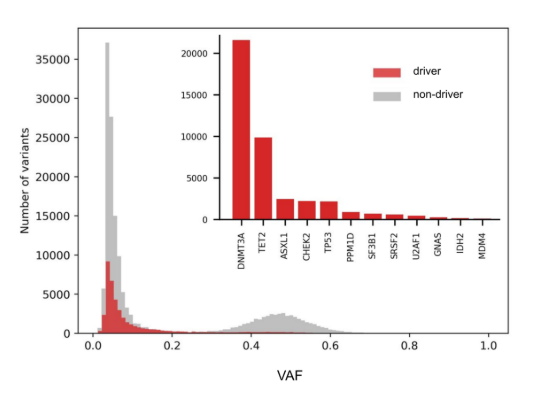
‼️ The final version of our article “Identification of Clonal Hematopoiesis Driver Mutations through In Silico Saturation Mutagenesis” is out in Cancer Discovery @CD_AACR: aacrjournals.org/cancerdiscover… Learn more about our findings in this thread 👇🏽

New in Cancer Landscapes from the July 1 issue— Comprehensive Genetic Profiling Reveals Frequent Alterations of Driver Genes on the X Chromosome in Extranodal NK/T-cell Lymphoma, by Yuta Ito et al. bit.ly/3VZ9ttn @kataoka_lab

🍭new pre-print! In collab. with @MissionBio , we developed single-cell Genotype-to-Phenotype (scG2P) for targeted co-capture of DNA & mRNA! We explore esophagus clonal mosaicism, defining clonal structure the phenotypic impact of somatic drivers biorxiv.org/content/10.110…

New from the May issue— Pan-Cancer Comparative and Integrative Analyses of Driver Alterations Using Japanese and International Genomic Databases, by @Sara_Horie, @YukiSaito_MDPhD, @kataoka_lab et al. bit.ly/3V0R0NP @NccriOfficial @NccCat
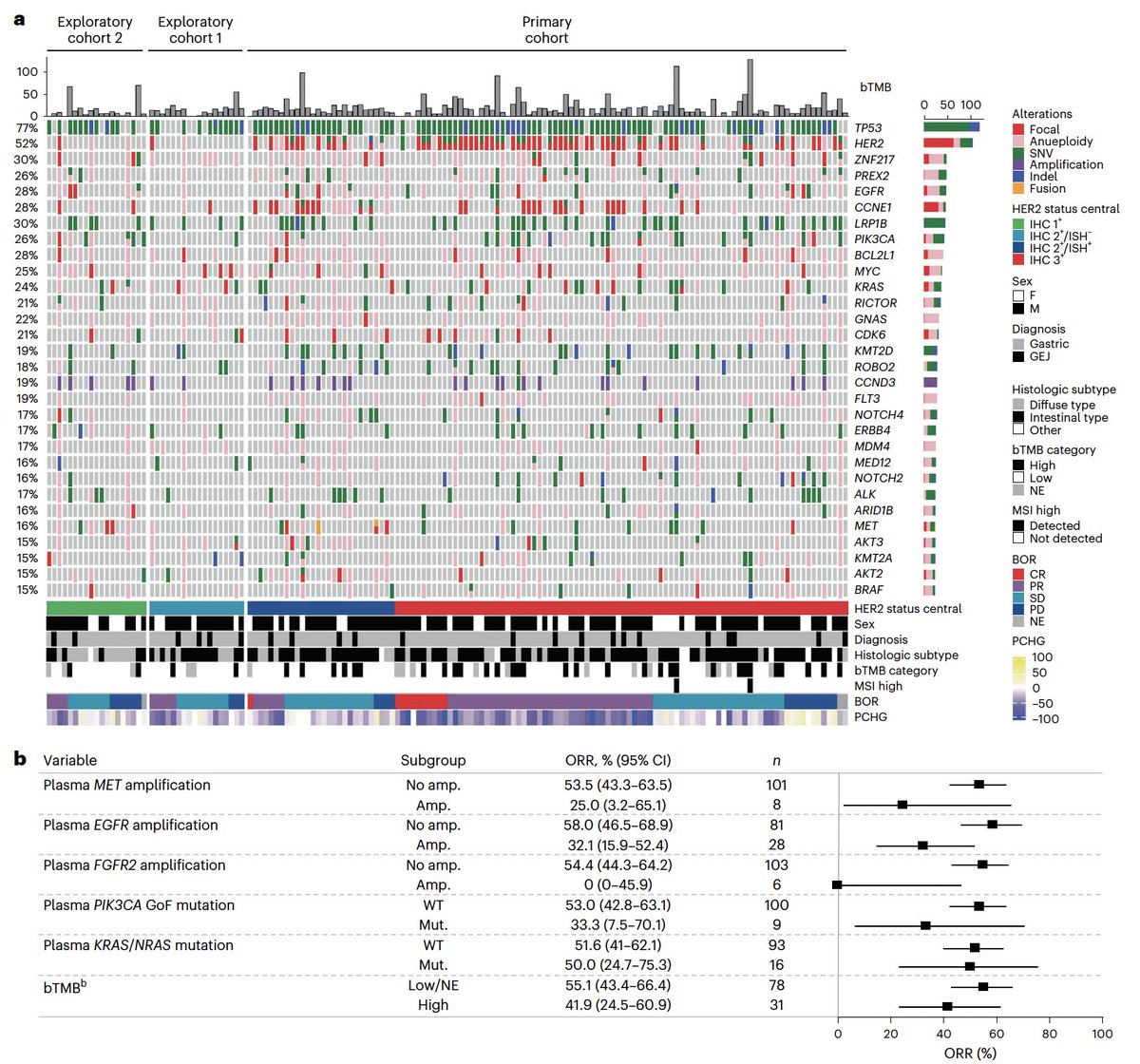
Trastuzumab deruxtecan in HER2+ gastric cancer @NatureMedicine doi.org/10.1038/s41591… 🔎exploratory biomarker analysis of DESTINY Gastric01 👉64% concordance btw HER2+ IHC & 🩸 👉MET, EGFR & FGFR2 GA: ORR⬇️ 👉HER2 mutations: ORR⬆️ 👉activity in Her2 low @myESMO
Understanding the complexity of p53 in a new era of tumor suppression p53が発見されて45年ですが、Cancer Cell誌に非常によくまとまったp53の総説が発表されました。 cell.com/cancer-cell/fu…
Now online in @CD_AACR: Identification of #ClonalHematopoiesis Driver Mutations through In Silico Saturation Mutagenesis - by Santiago Demajo, Joan Enric Ramis-Zaldivar, @abel_gonzalezp, @nlbigas and colleagues doi.org/10.1158/2159-8… @bbglab @irbbarcelona @CIBERONC
A paper in @Nature reports on the ability of AlphaFold 3 to produce accurate predictions of the structures of proteins interacting with other biological molecules. AlphaFold 3 demonstrates substantially better accuracy over previous specialist tools. go.nature.com/44CAnev
Pan-cancer comparative and integrative analyses of driver alterations using Japanese and international genomic databases [Jan 26, 2024] @Sara_Horie @YukiSaito_MDPhD @kataoka_lab et al. @CD_AACR bit.ly/490njke #PrecisionMedicine #globonc @NccriOfficial @NccCat
United States Trends
- 1. Indiana 38,3 B posts
- 2. Ohio State 26,7 B posts
- 3. Caleb Downs 5.693 posts
- 4. $AROK 6.401 posts
- 5. Wayne 120 B posts
- 6. Howard 22,4 B posts
- 7. #daddychill 2.536 posts
- 8. Gus Johnson 1.188 posts
- 9. Buckeyes 8.053 posts
- 10. Ryan Day 3.728 posts
- 11. UMass 3.672 posts
- 12. #iufb 4.745 posts
- 13. Hoosiers 7.749 posts
- 14. Tottenham 52,1 B posts
- 15. Surgeon General 123 B posts
- 16. Man City 44,5 B posts
- 17. Spurs 39,6 B posts
- 18. DJ Lagway 1.897 posts
- 19. #GoBucks 4.665 posts
- 20. Chip Kelly N/A
Who to follow
-
 Keisuke Kataoka Laboratory(片岡 圭亮 研究室)
Keisuke Kataoka Laboratory(片岡 圭亮 研究室)
@kataoka_lab -
 YukinoriOkada
YukinoriOkada
@okada_yukinori -
 Takashi Kohno@Natl Cancer Ctr, Japan
Takashi Kohno@Natl Cancer Ctr, Japan
@TakashiKohno2 -
 白石 友一
白石 友一
@friend1ws -
 河野暉
河野暉
@hikarundesuyo -
 Takahiro Maeda
Takahiro Maeda
@TMHemOnc -
 Shinichi Namba
Shinichi Namba
@NambaShinichi -
 みなみ
みなみ
@rmhtgy373south -
 Seiichi Mori
Seiichi Mori
@seiichi_mori -
 Monitarou
Monitarou
@nikuwoyakuman -
 Takahiro Kamiya
Takahiro Kamiya
@Know_the__Truth -
 YotarOchi
YotarOchi
@yota_ochi -
 Masato Akiyama
Masato Akiyama
@akiyama_mst -
 たくみ
たくみ
@changumu1 -
 Hiro Suzuki (Hiromichi Suzuki)
Hiro Suzuki (Hiromichi Suzuki)
@HiromicsSuzuki
Something went wrong.
Something went wrong.