Subhan Hadi
@SubhanhadiPostdoctoral at SANKEN, Osaka University
Similar User

@NasuLab_IBC

@sammyhschan

@jiayihh

@muridjalanan

@frastinau

@gatauakupusingg
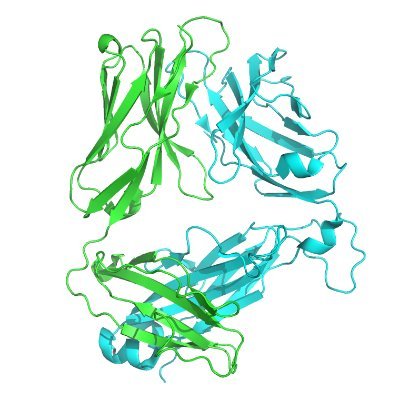
Autonomous multicolor bioluminescence imaging in bacteria, mammalian, and plant hosts | PNAS pnas.org/doi/10.1073/pn…
Language Models for Molecular Dynamics • MDLMs (Molecular Dynamics Language Models) bridge language model capabilities with the physical accuracy of molecular dynamics (MD), enabling efficient generation of MD trajectories. • MDLMs predict protein dynamics using just 5% of a…

Happy Friday everyone! Here are some cool Arabidopsis mini trees🌴 Arabidopsis arenosa hypocotyls can undergo extensive secondary growth and the rosettes that develop on top make them look like palm trees 🤯I wonder if this can be engineered in thaliana..


Exploration of Cryptic Pockets Using Enhanced Sampling Along Normal Modes: A Case Study of KRAS G12D #MolecularDynamics pubs.acs.org/doi/10.1021/ac… @VithaniNeha @ABalaeff @OpenEyeSoftware #JCIM Vol64 Issue21 #compchem
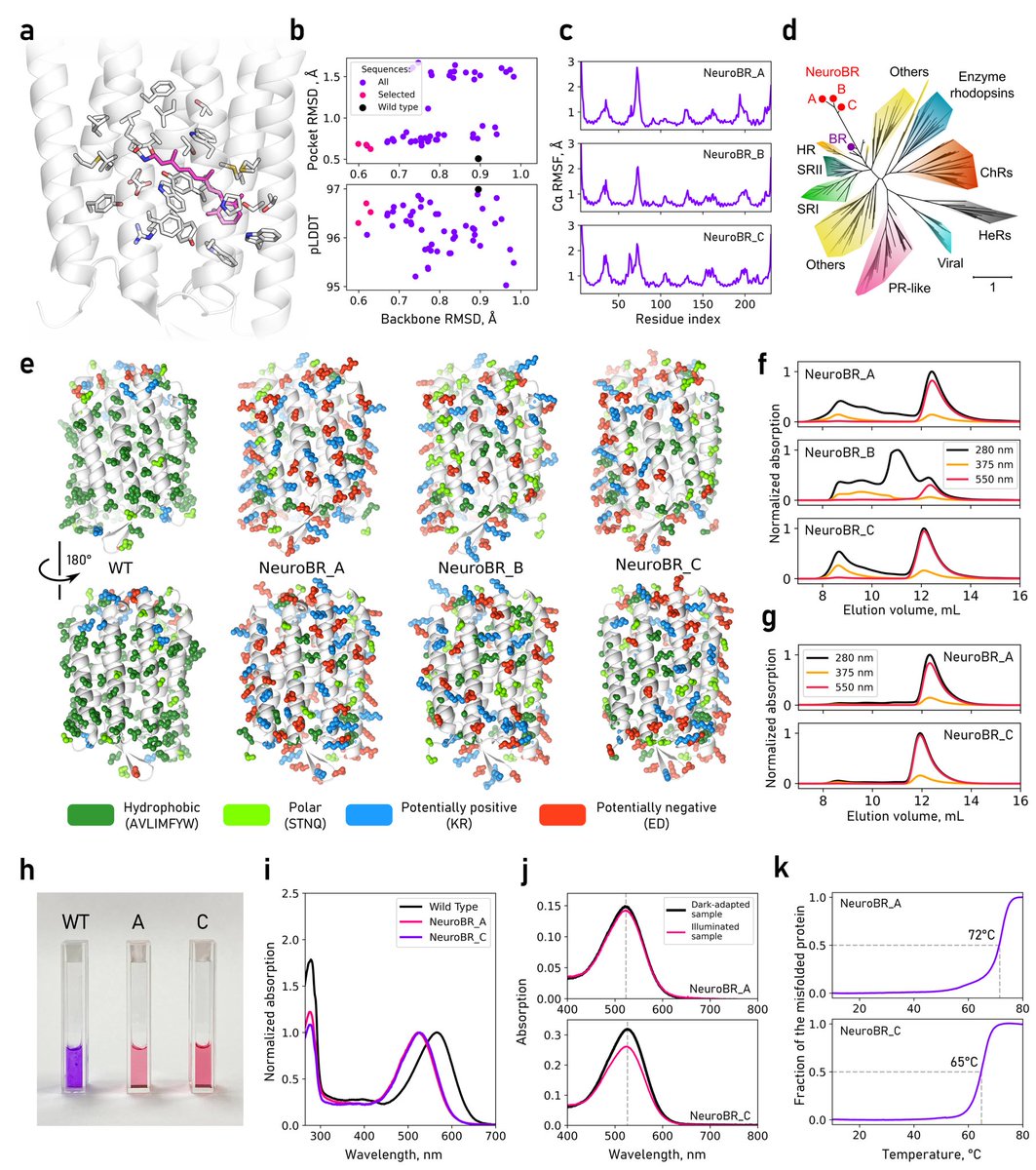
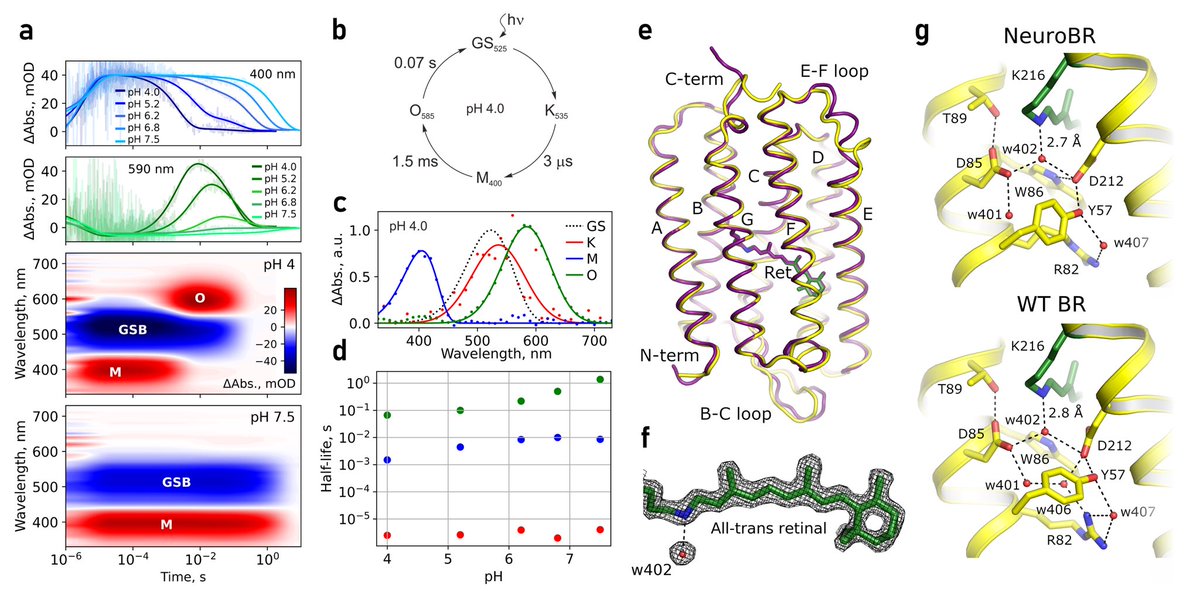
We engineered membrane protein bacteriorhodopsin to be soluble. It binds retinal and can photocycle! X-ray structure reveals conserved binding pocket with 0.8 Å all atom RMSD to WT BR. All this became possible due to hard work by Andrey Nikolaev and the team 🙏 #proteindesign
This is great 👍 Now we only need a proper benchmark to see that none of these models actually work.
Thrilled to announce Boltz-1, the first open-source and commercially available model to achieve AlphaFold3-level accuracy on biomolecular structure prediction! An exciting collaboration with @jeremyWohlwend, @pas_saro and an amazing team at MIT and Genesis Therapeutics. A thread!
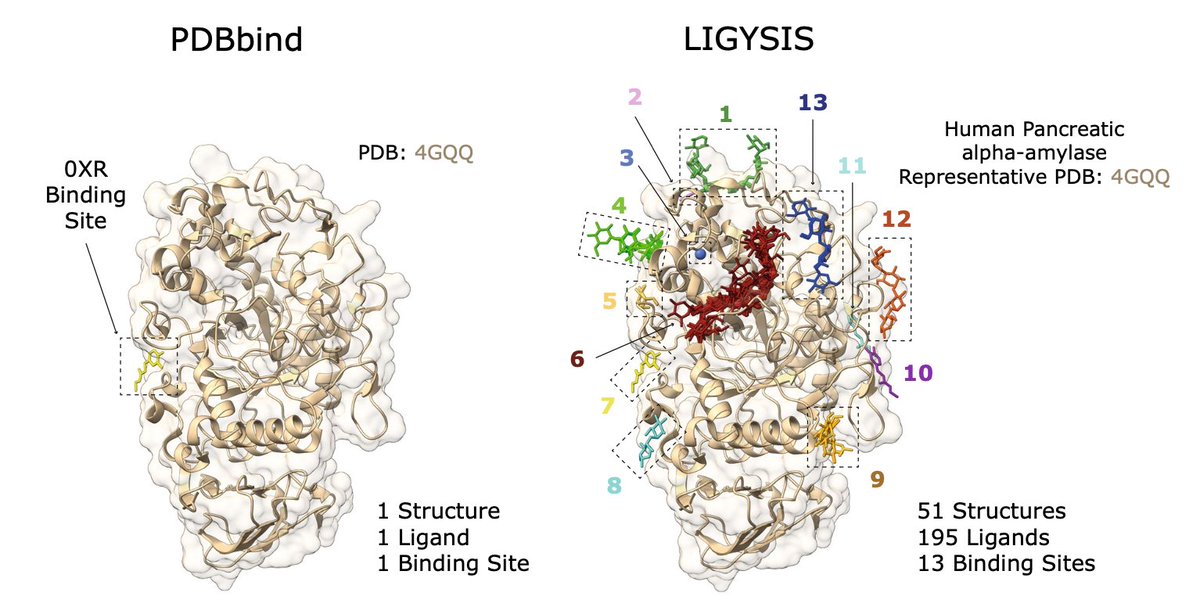
Comparative Evaluation of Methods for the Prediction of Protein–Ligand Binding Sites @jcheminf 1. This study benchmarks 13 state-of-the-art protein–ligand binding site prediction methods across three decades, introducing the new LIGYSIS dataset with 30,000 biologically relevant…
#HighlightOfTheWeek Cosolvent molecular dynamics are an increasingly popular form of MD simulations where small molecule cosolvents are added to the system, helping to identify cryptic and allosteric pockets in proteins. #compchem pubs.acs.org/doi/10.1021/ac…
New Article: "The complete genome assembly of Nicotiana benthamiana reveals the genetic and epigenetic landscape of centromeres" rdcu.be/d0h6Q ...revealing the structure, epigenetic landscape and evolutionary dynamics of its centromeres following allotetraploidization.

The vacuolar K+/H+ exchangers and calmodulin-like CML18 constitute a pH-sensing module that regulates K+ status in Arabidopsis | Science Advances science.org/doi/10.1126/sc…
A new Science study presents “Evo”—a machine learning model capable of decoding and designing DNA, RNA, and protein sequences, from molecular to genome scale, with unparalleled accuracy. Evo’s ability to predict, generate, and engineer entire genomic sequences could change the…

A machine learning system that simulates proteins with more than 10,000 atoms with quantum accuracy. nature.com/articles/s4158…




De novo design of triosephosphate isomerases using generative language models 1. This study demonstrates the power of large language models (LLMs) for the de novo design of functional enzymes, specifically triosephosphate isomerases (TIMs), by using two generative models,…

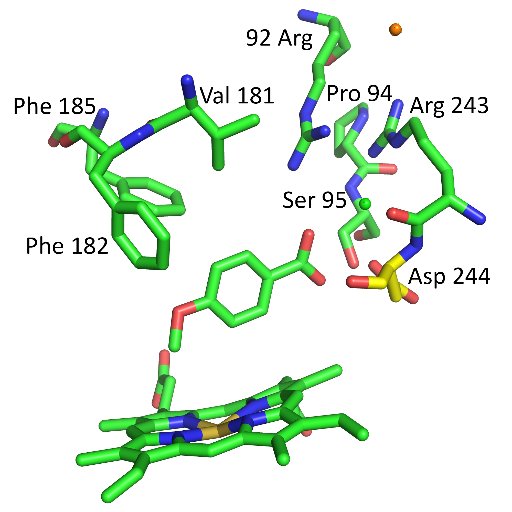
PDBe tools for an in-depth analysis of small molecules in the Protein Data Bank 1. PDBe has developed new tools to facilitate the analysis of small molecules in the PDB, enhancing data accessibility and insight into ligand-macromolecule interactions. The tools—PDBe CCDUtils,…

Evolution-assisted engineering of E. coli enables growth on formic acid at ambient CO2 via the Serine Threonine Cycle "synthetic C1-utilization pathways are engineered into E. coli" sciencedirect.com/science/articl…

Leveraging Fluorescence Lifetime Imaging Microscopy & phasor analysis for precise measurement of pH of insulin secretory granules with subcellular resolution. @KateWhiteLab pubs.acs.org/doi/10.1021/ac…
AlphaFold can predict protein, DNA, or RNA complexes.. BUT is it easy to determine the reliable interactions?🤔 ➡️ AlphaBridge will identify the most confident interactions, making your analysis easier! 🌐Try it here: alpha-bridge.eu 👀Read more: biorxiv.org/content/10.110…

Binary vector copy number engineering improves Agrobacterium-mediated transformation go.nature.com/3CdewQS

Free scientific illustrations for biologists! 😍 @NIH has released a library of 500+ free scientific illustrations to create figures, presentations, and illustrations! all freely available in the public domain. Retweet and spread the message! bioart.niaid.nih.gov

United States Trends
- 1. $LFDOG 5.341 posts
- 2. #SmallBusinessSaturday 4.350 posts
- 3. $CUTO 7.037 posts
- 4. Las Palmas 40,9 B posts
- 5. Lando 44,2 B posts
- 6. Real ID 7.103 posts
- 7. #Caturday 5.959 posts
- 8. Go Bucks 1.830 posts
- 9. Gameday 48,7 B posts
- 10. Go Blue 12,7 B posts
- 11. #ShopSmall 3.015 posts
- 12. #SaturdayVibes 6.065 posts
- 13. Trudeau 191 B posts
- 14. Bournemouth 7.888 posts
- 15. Grok 53,8 B posts
- 16. Good Saturday 33,2 B posts
- 17. Zelensky 133 B posts
- 18. Barcelona 117 B posts
- 19. #TheGame N/A
- 20. Fermin 10,6 B posts
Something went wrong.
Something went wrong.