Jonathan Mitchel
@J_E_MitchelPhD candidate at Harvard-MIT HST @mit_hst. Studying cells with computers. Computational biology | Single-cell genomics | Coffee |
Similar User

@kaessmannlab

@mrjeffhsiao

@OwenKLeddy

@lmwebr

@seankenneths

@blake_lash

@PratikWakode13
Stem cell gene therapy can be used to treat diverse genetic diseases. How does a person's disease background affect gene-edited stem cell activity? A pleasure writing this @NatureNV piece with @bloodgenes summarizing a fantastic study by Calabria & co nature.com/articles/d4158…
I am excited to share that I will be starting a lab at @EmoryUniversity this September. We will aim to understand evolution by studying genomes of things ranging from viruses to humans. Looking for talented students and postdocs to do computational genomics. Get in touch! 1/2
1/10 [My first Twitter/X post] Our preprint about the joint statistical analysis of the Undiagnosed Diseases Network patients is finally out: biorxiv.org/content/10.110…
Very excited to share that I recently started my lab in @CompOncMSK at @MSKCancerCenter! Looking for postdocs and graduate students interested in cell fate decisions/cancer evolution/computational single-cell genomics. Come join us in NYC! Spread the word & please #RT. Thanks!
I’ve learned about expectation-maximization (EM) algorithms many times in classes and in papers, but it has always been unclear to me how to derive an EM algorithm to solve a new problem. So I spent some time putting together this tutorial. Enjoy! teng-gao.github.io/blog/2022/ems/
Preprint alert📣📣Excited to share our new method SCENT to create accurate enhancer-gene map from #singlecell #multiome data with @soumya_boston! SCENT using multiome technology is very powerful in defining disease causal variants & genes in #GWAS #ASHG22 medrxiv.org/content/10.110…

I think about this question a lot. The most inspiring answer I've heard (from @KharchenkoLab) is that the conceptual advance introduced in a new method is often more important than the implementation itself, and will live on long after the original code is obsolete.
Question for my computational methodology folks, what keeps you going developing computational methods knowing that some better, faster, more creative methods will be out, sooner or later? #thoughtsOftheday
Recommended: thought-provoking challenge to our current mainstream post-GWAS approaches to find causative variants, led by Shamil Sunyaev's group @HarvardDBMI The missing link between genetic association and regulatory function medrxiv.org/content/10.110…
Excited to share our paper describing Numbat, a haplotype-aware CNV caller for scRNA-seq, out today on @NatureBiotech! nature.com/articles/s4158… (1/5)
@EBiederstedt introducing the Cell Annotation Platform at celltype.info highlighting the many people contributing to this exciting effort spearheaded by @KharchenkoLab

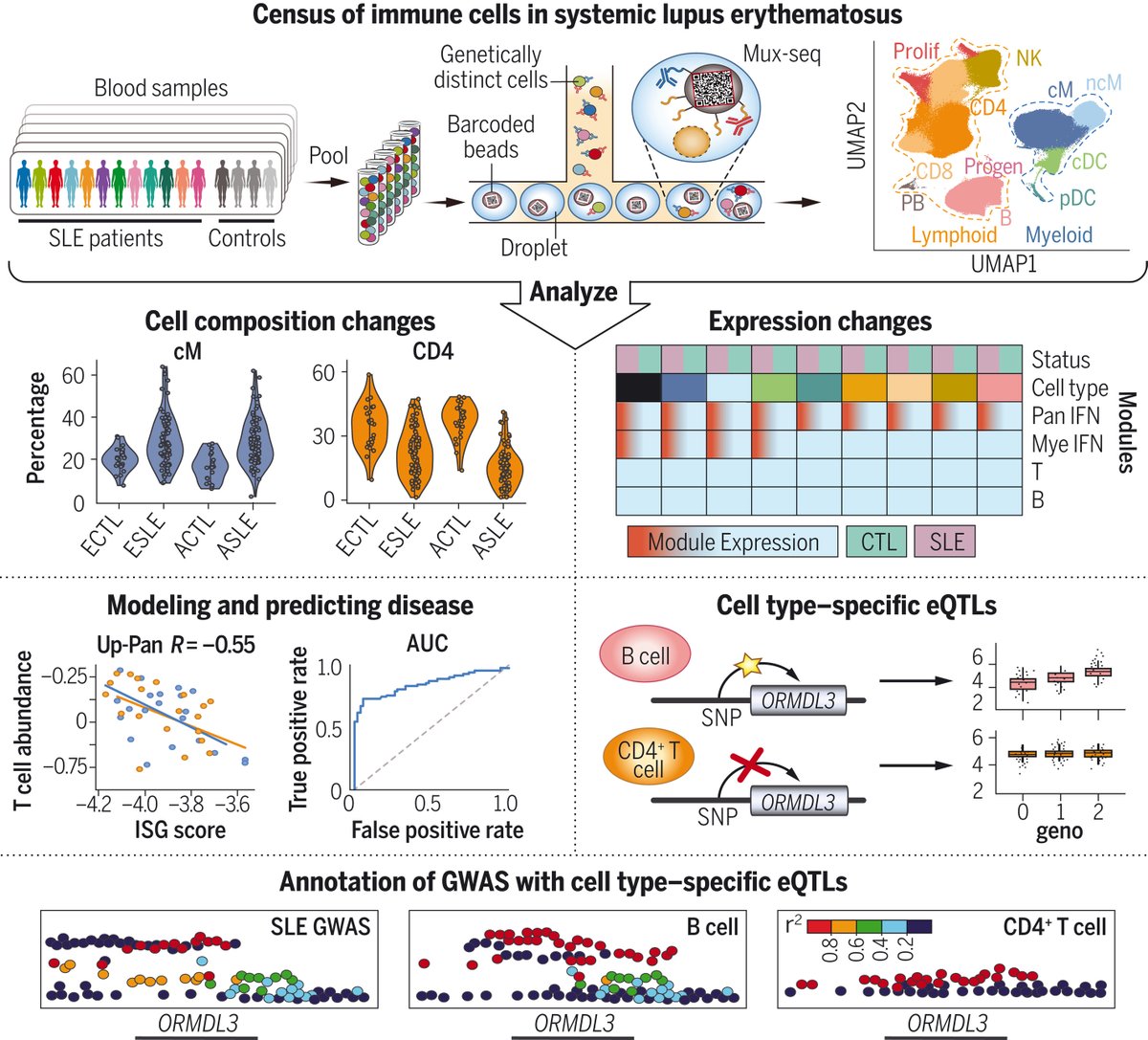
In 2018, we developed mux-seq and demuxlet to enable population-scale single-cell sequencing. Today, excited to share our paper in @ScienceMagazine where we deployed mux-seq to profile 1.2M immune cells from 162 lupus patients and 99 healthy controls: science.org/doi/full/10.11…. 1/n
Comparison of two or more scRNA-seq panels is becoming increasingly common. Based on our experience we've put together Cacoa - an R package to carry out statistical tests, exploration, and visualization of scRNA-seq panel contrasts. 1/n biorxiv.org/content/10.110…
T-cells provide a natural orthogonal test of clonality using the TCR sequence, so we created a new method to detect TCR sequence from 10x 3’ scRNA-seq: TREK-seq! (previously needed 10x 5' for TCR detection) – assist from @shaleklab & Love labs @MIT!
United States Trends
- 1. Wayne 119 B posts
- 2. Neil 28,9 B posts
- 3. Saka 62,3 B posts
- 4. Red Cross 11,5 B posts
- 5. Surgeon General 81,4 B posts
- 6. Gameday 34,6 B posts
- 7. Arsenal 116 B posts
- 8. #UFCMacau 35,6 B posts
- 9. #Caturday 6.908 posts
- 10. #saturdaymorning 5.121 posts
- 11. Odegaard 11,7 B posts
- 12. #ARSNFO 8.208 posts
- 13. #Arcane 522 B posts
- 14. Good Saturday 35,5 B posts
- 15. Petr Yan 6.599 posts
- 16. Partey 10,3 B posts
- 17. Weezy 4.395 posts
- 18. Buckeyes 3.401 posts
- 19. Enzo 79,4 B posts
- 20. Figgy 3.834 posts
Something went wrong.
Something went wrong.