Yasuhiro Arimura
@ArmrYshrElucidating the structural basis of biological events on chromosomes | chromatin, Xenopus egg extract, MagIC-cryo-EM | Assistant Professor at @HutchBasicSci
Similar User

@kazu_maeshima

@Nucleosome_Bot

@FachinettiLab

@FukayaLab

@TjianDarzacq

@robert_ife

@TomoKujira

@Jan_Soro

@RobKlose

@Hake_Chromatin

@YuanHeLab

@VashKurdistani

@ZierLab

@tatsuofukagawa1

@SG_Swygert
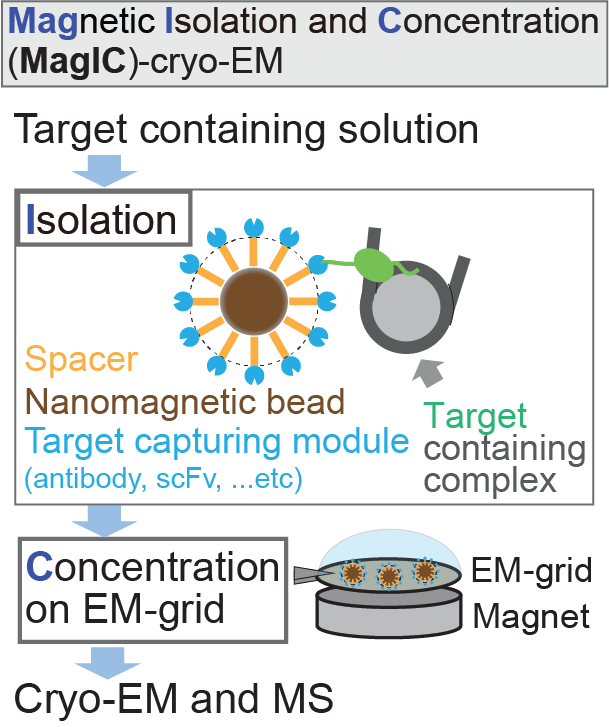
Magnetic Isolation and Concentration (MagIC)-cryo-EM is a new method to enable direct structural analysis of targets captured on nanomagnetic beads, which requires only 0.0005 mg/mL of targets in 100 µL crude solution to make 10 cryo-EM grids. I will explain the magic behind it!
Like astronauts exploring a planet, no stone is left unturned in the @GiuntaLab when it comes to human DNA 🧬Our computational simulations show that centromere sequences are intrinsically prone to complex and unstable secondary structures correlated with chromosome missegregation
🚨 New from Chittoor & @Simona_Giunta 📰Comparative analysis of predicted DNA secondary structures infers complex human centromere topology cell.com/ajhg/fulltext/…
Excited that our work on transcription-coupled DNA repair is now published @CellCellPress! We recapitulated TC-NER in Xenopus egg extract and identified STK19 as a core repair factor that couples stalled Pol II to TFIIH. 🧬🐸 ➡️ cell.com/cell/fulltext/…
最も尊敬する女性研究者の訃報。。私がモデル植物シロイヌナズナの発生遺伝学者を志すきっかけであり、ロールモデルでもある憧れの偉大な研究者。RIP, Joanne, you are my hero and the reason why I became a plant developmental biologist...🌱
Please see our paper in Nature on read-write mechanisms of H2AK119 ubiquitination by Polycomb repressive complex I. Congrats to the whole team, especially Victoria and huge thanks to our collaborator @valendraica ! Thank you @TheMarkFdn for your support! rdcu.be/dZ5HZ
This week's lesson: You never know until you try. You might discover a hidden talent. (DH5a sometimes provides the best protein expression🙃)

hey #teamtomo if you are looking to add to your toolkit for molecular identification inside cells, our preprint is now out!
ExoSloNano: Multi-Modal Nanogold Tags for identi-fication of Macromolecules in Live Cells & Cryo-Electron Tomograms biorxiv.org/cgi/content/sh… #biorxiv_biophys
I have written a concise review on chromatin transcription as part of the @JMolBiol special issue on transcription elongation. Check it out! authors.elsevier.com/sd/article/S00…

Our new paper is out @NatureCellBio🥳It shows how major epigenetic OFF mark, H3K27me3, is newly established at the beginning of life. This work was mainly carried out by a talented PhD student, Masahiro Matsuwaka👏 nature.com/articles/s4155… rdcu.be/dYJf Short summary↓↓

Great faculty position in VIDD @Fredhutch open-strong support from the Bezos family included! Please apply and/or RT!
Join us in accelerating groundbreaking discoveries in #infectiousdiseases and #immunology! Fred Hutch is seeking a scientific leader for the Bezos Family Distinguished Scholar in Viruses and Vaccines faculty position. Learn more and apply: bit.ly/488PkH4

BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”

The @WeintraubAward recognizes the outstanding achievements of grad students in biology and was established to honor the bold, creative, pioneering spirit embodied by Dr. Hal Weintraub. Nominations for 2025 are now open!
Nominations for the 2025 Weintraub Award are now open! Celebrate the achievements of an outstanding graduate student in biology by nominating them here: fredhutch.org/en/research/di…
Please nominate grad students for the @WeintraubAward!
The @WeintraubAward recognizes the outstanding achievements of grad students in biology and was established to honor the bold, creative, pioneering spirit embodied by Dr. Hal Weintraub. Nominations for 2025 are now open!
What is the 3D structure of chromatin condensed by the Polycomb repressive complex 1 (PRC1)? We determined it using #cryoET. Led by @MichaUckelmann from the @DavidovichLab in collaboration with @demarco_lab @EMBLAustralia @MonashBDI @CryoEM_Monash biorxiv.org/content/10.110…
Check out this news story on our paper nature.com/articles/s4159…
New findings from @InLiuOfBulkExps provide insight into a master regulator of gene expression, and could open up new avenues for Rett syndrome therapies. #RockefellerScience rockefeller.edu/news/36409-new…
Particularly excited to share our latest work, where we visualize how myosin motor forces alter the structure of actin. We discovered a spiral form of the filament which can be recognized by force-sensitive actin binding proteins. biorxiv.org/content/10.110…
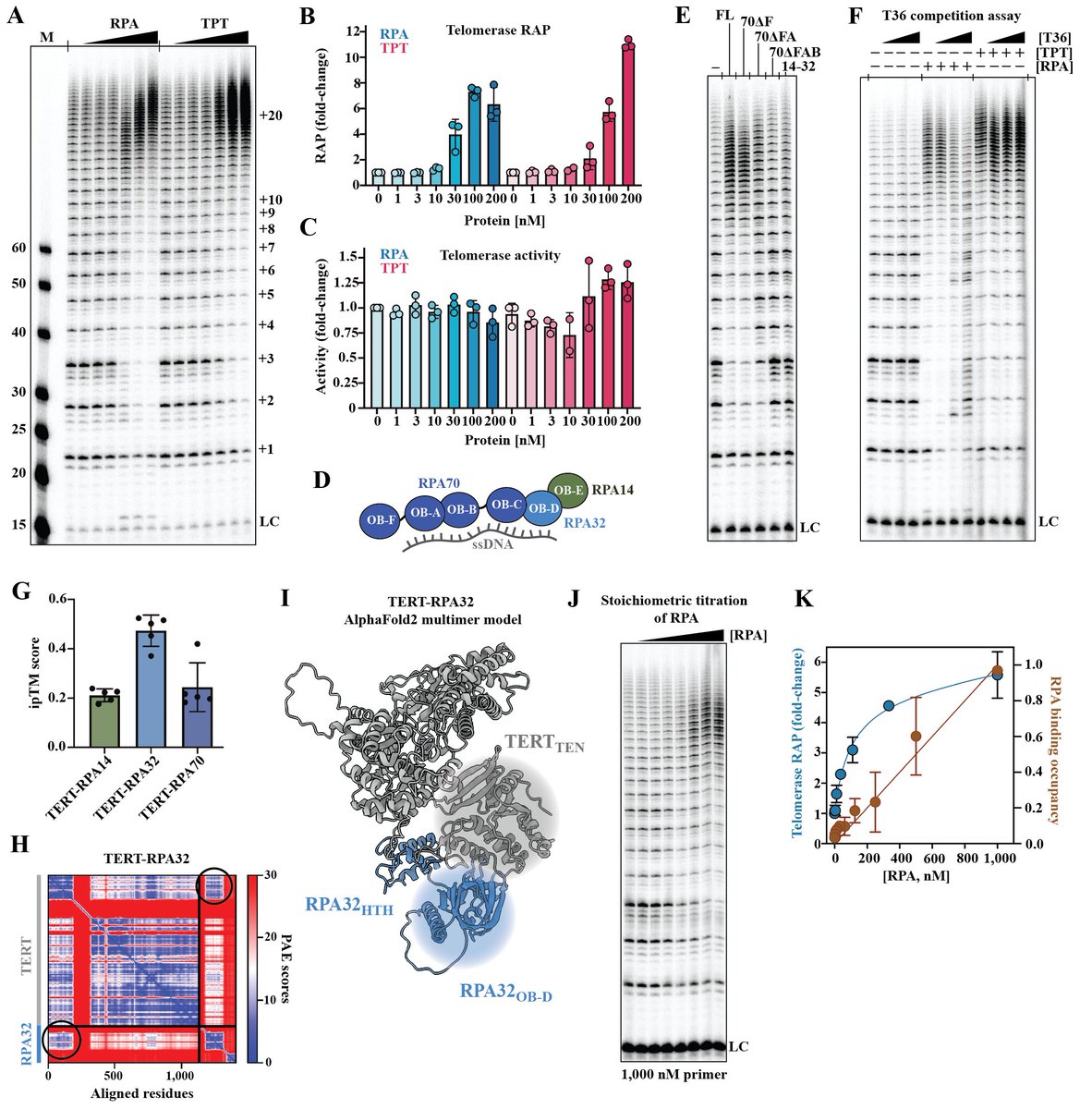
1/7 Our latest preprint is out! 🚨 @Sour_Agrawal , @xiuhua_linwisc , @viveksusvirkar et al. discover human RPA is a critical telomerase processivity factor essential for telomere maintenance. This reshapes our understanding of human telomerase regulation. biorxiv.org/content/10.110…
Xenopus Resources and Emerging Technologies (XRET) Meeting: October 11-14, 2024 at the Marine Biological Laboratory, Woods Hole, MA USA xenbase.org/xenbase/doNews… #science #biology #devbio #xenopus #frogs

Please RP! We are hiring a postdoc to explore how eukaryotic chromatin evolved from Archaea! This position is funded for 3 years by #HFSP. jrecin.jst.go.jp/seek/SeekJorDe…
ポスドクの公募を出しました! HFSPの支援のもと、アーキアクロマチンの研究をしてくださるポスドクの方を募集します。良い応募者の方がいたらセミナー形式の研究発表をお願いする予定です(現地またはオンライン、現地の場合は交通費支給あり)。 kyoto-u.ac.jp/sites/default/…
New preprint combining chromatin simulations with CNNs to quantitatively interpret RICC-seq data! So excited about this work led by @ArianaClerkin with major contributions from @npagane and help from Devany West, in collaboration with @ASpakowitz👇 biorxiv.org/content/10.110…
We are excited to present a flexible modeling and simulation framework that boosts our ability to draw inferences about mesoscale chromatin structure using RICC-seq data. biorxiv.org/content/10.110… (1/4)🧵
Our review on using single-molecule correlative force and fluorescence microscopy to study protein-DNA interactions is out! Honored to share our perspective on this fast-growing field. Thanks to @AnnualReviews for the open access volume 👍🏼 doi.org/10.1146/annure… @BadAtCloning

United States Trends
- 1. #ArcaneSeason2 145 B posts
- 2. #UFCMacau 16,5 B posts
- 3. Jayce 64,4 B posts
- 4. Ekko 59,2 B posts
- 5. Wayne 89,2 B posts
- 6. SEVENTEEN 1,36 Mn posts
- 7. MADDIE 20,1 B posts
- 8. Jinx 190 B posts
- 9. Good Saturday 21,6 B posts
- 10. Ulberg 2.111 posts
- 11. Shi Ming 1.830 posts
- 12. #saturdaymorning 2.853 posts
- 13. #SaturdayVibes 4.213 posts
- 14. woozi 294 B posts
- 15. #jayvik 62,4 B posts
- 16. BIGBANG 411 B posts
- 17. Wang Cong N/A
- 18. Volkan 3.884 posts
- 19. Jungkook 539 B posts
- 20. Feng 6.866 posts
Who to follow
-
 Kazuhiro Maeshima
Kazuhiro Maeshima
@kazu_maeshima -
 Nucleosome papers 📚 NucPosDB database
Nucleosome papers 📚 NucPosDB database
@Nucleosome_Bot -
 Daniele Fachinetti
Daniele Fachinetti
@FachinettiLab -
 Takashi Fukaya
Takashi Fukaya
@FukayaLab -
 Tjian + Darzacq Group
Tjian + Darzacq Group
@TjianDarzacq -
 Robert Schneider
Robert Schneider
@robert_ife -
 Tomoya Kujirai
Tomoya Kujirai
@TomoKujira -
 Jan Soroczynski
Jan Soroczynski
@Jan_Soro -
 Rob Klose
Rob Klose
@RobKlose -
 Sandra Hake
Sandra Hake
@Hake_Chromatin -
 The He Lab
The He Lab
@YuanHeLab -
 Siavash Kurdistani
Siavash Kurdistani
@VashKurdistani -
 Christian Zierhut
Christian Zierhut
@ZierLab -
 Tatsuo Fukagawa
Tatsuo Fukagawa
@tatsuofukagawa1 -
 Sarah G. Swygert
Sarah G. Swygert
@SG_Swygert
Something went wrong.
Something went wrong.